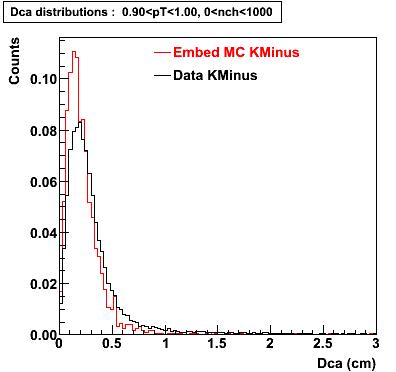
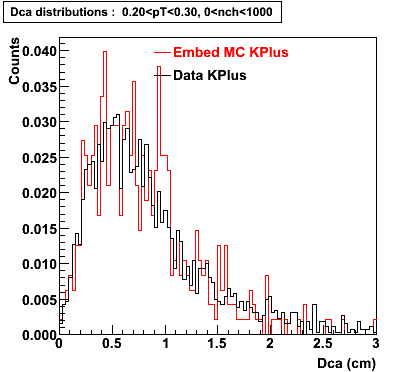
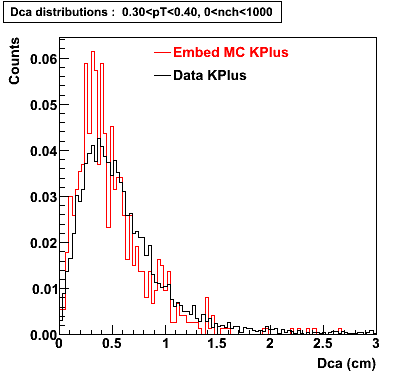
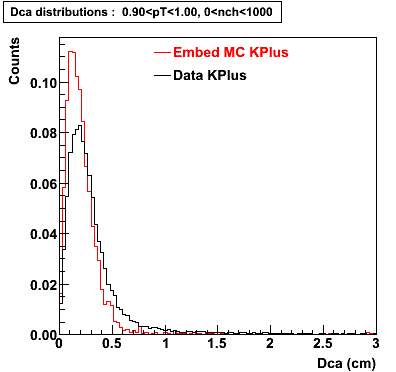
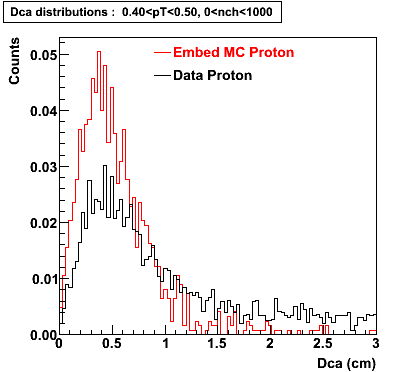
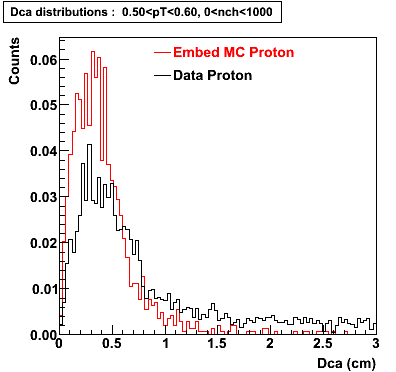
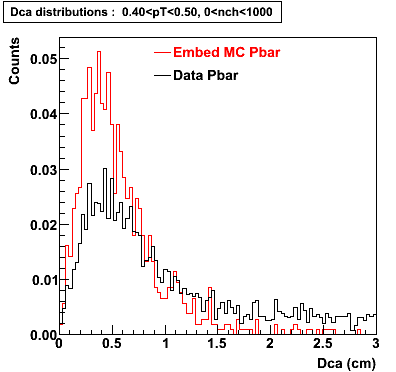
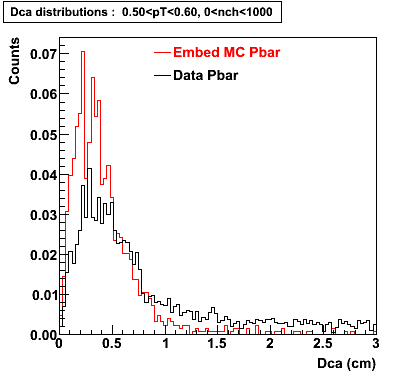
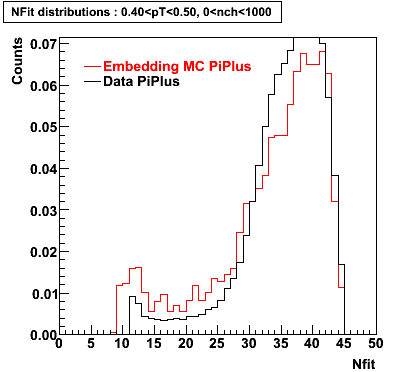
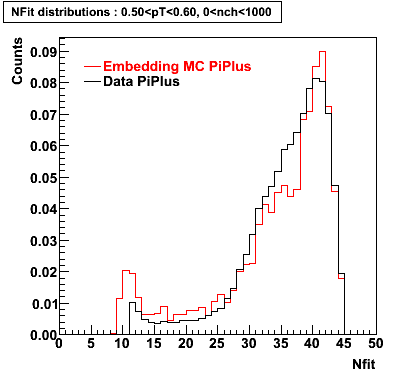
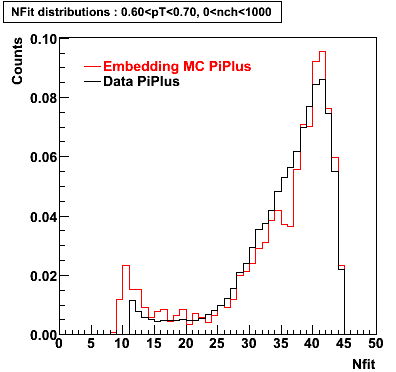
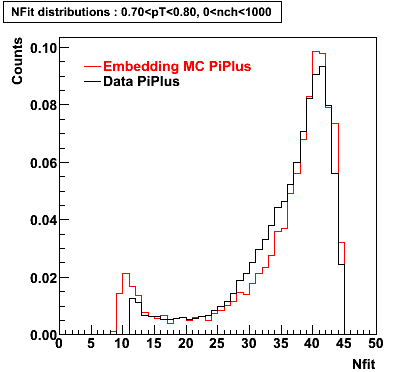
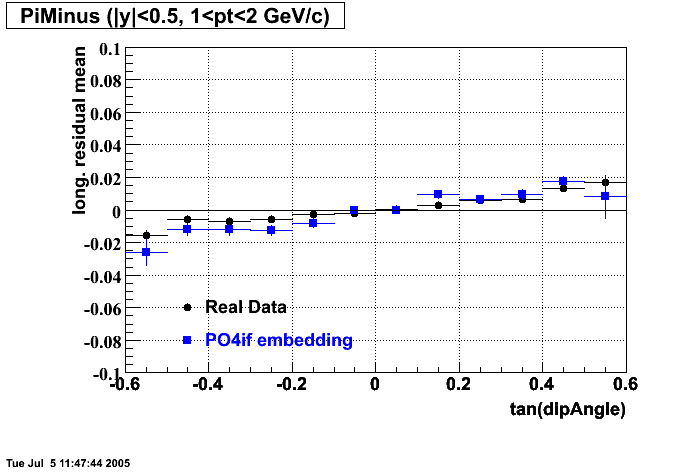
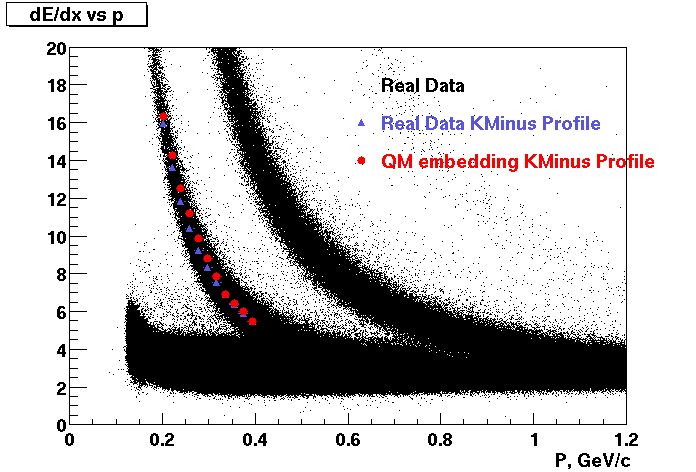
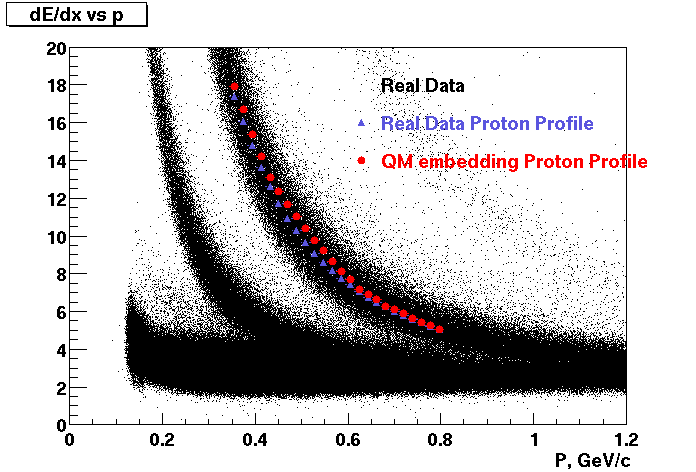
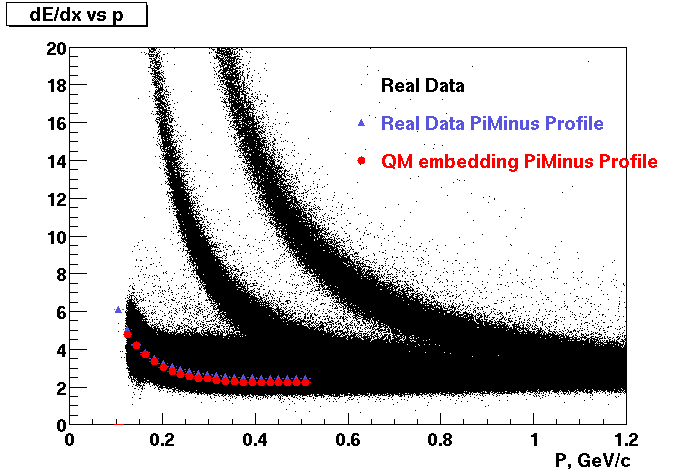
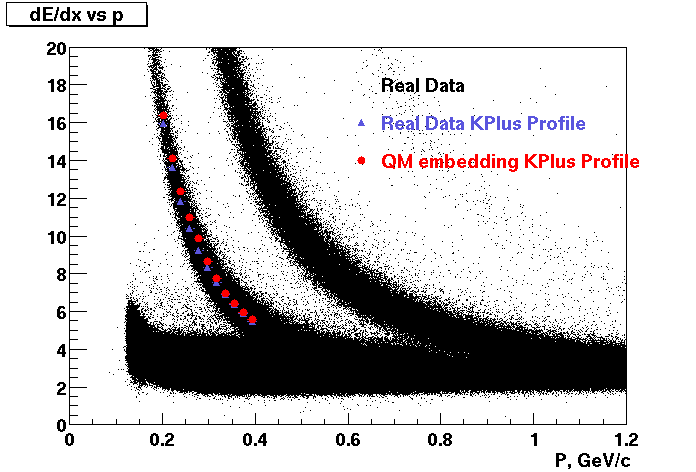
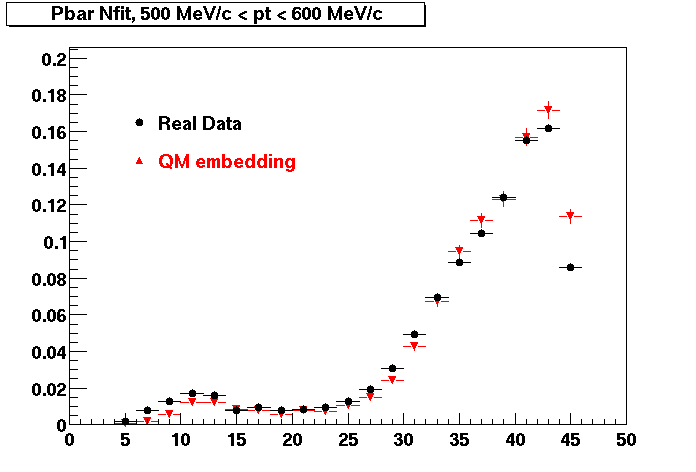
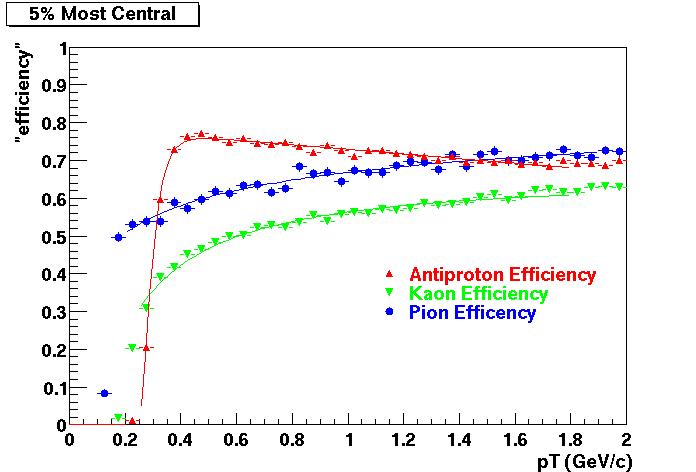
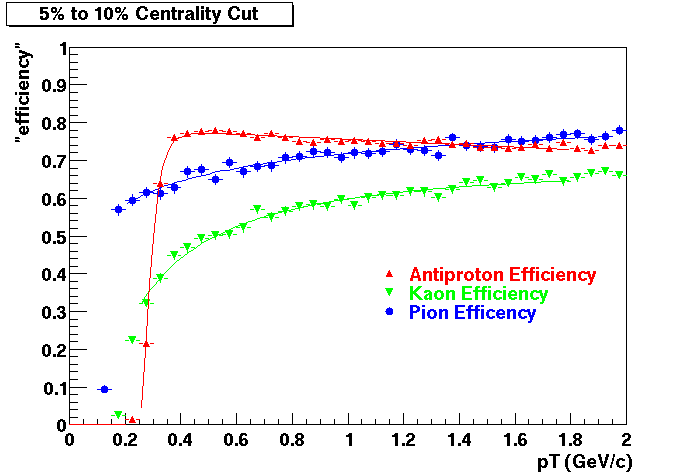
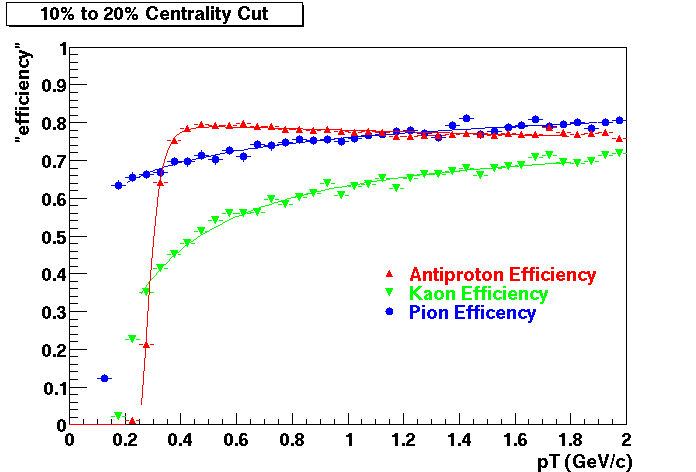
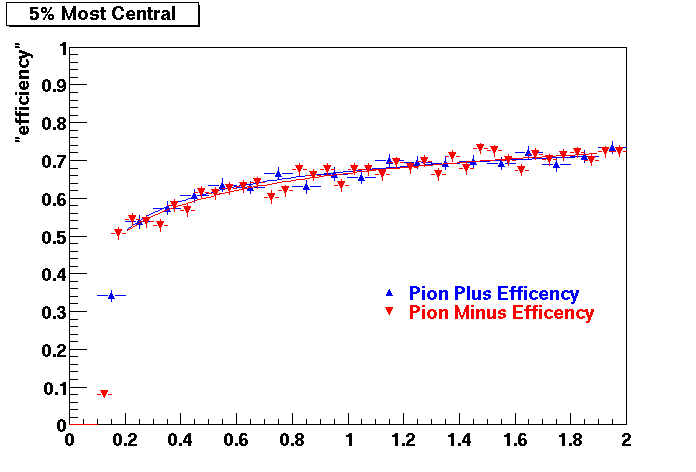
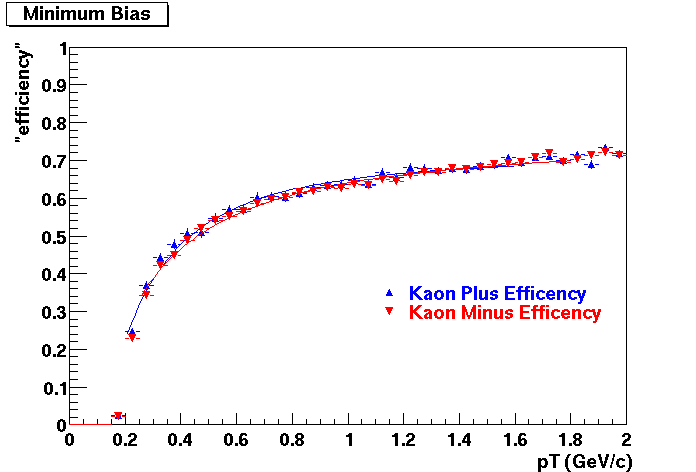
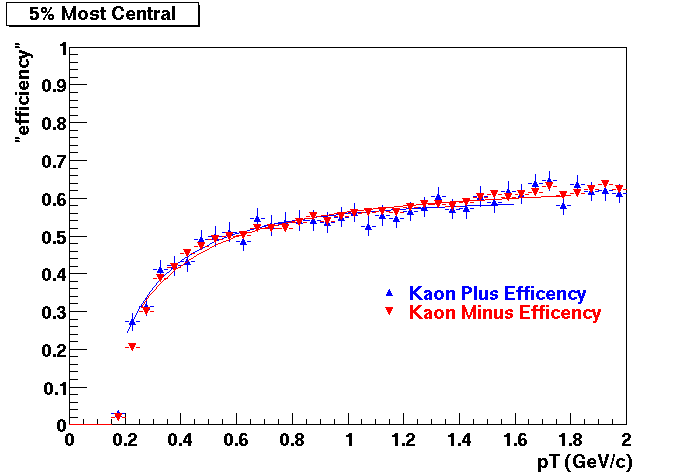
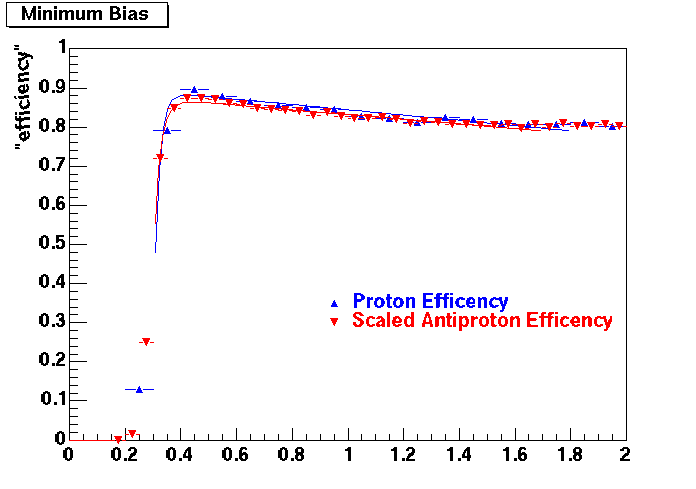
Embedding
Welcome to the STAR Embedding Pages!
Embedding data are generally used in STAR experiments for detector acceptance & reconstruction efficiency study. In general, the efficiency depends on running conditions, particle types, particle kinematics, and offline software versions. In principle, each physics analysis will need to formulate its own embedding requests by providing all the above relevant information. In STAR Collaboration, the embedding team is assigned to process those embedding requests and provide these embedding data, you can find out how the embedding team works in the Embedding structure page.
Over the past years, lots of embedding requests from different PWG's have been processed by the embedding team, please find the list in STAR Simulations Requests interface. If you want to look at some of these data, but do not know where these data are stored, please go to this page. If you can not find similar embedding request, you need to formulate your own embedding request for your particular study, please go to this page for more information about how to formulate an embedding request.
Please subscribe to the embedding mailing list if you are interested with embedding discussion: Starembd-l@lists.bnl.gov
And please join our weekly embedding meeting : https://drupal.star.bnl.gov/STAR/node/65992
Finding existing embedding data
Embedding data were produced for each embedding request in the STAR Simulations Request page.
Normally, they will be stored in RCF NFS disks for a while for end users to do their analysis.
However, NFS disk space is very limited, and we have new requests constantly, the data will be finally moved
from disks to HPSS for permanent storage on tape, but they can be restaged to disk for analysis later.
In order to find the existing embedding either on disks or in HPSS. Please follow the procedures below:
1) Find the request ID of a particular request that you are interested in, in the STAR Simulations Request page.
You can use the "Search" box at the top right of this page. Once you find the entry, look at the 'Request History' tab for more information, usually the original NFS data directory (when the data was first produced) can be found there.
2) Currently, the RCF NFS disks for embedding data are /star/data105, /star/embed and /star/data18.
For the data directories of each request, please logon to RCF, look at '/star/data105', '/star/embed' or '/star/data18' to see whether the following directories exist:
/star/data105/embedding/${Trigger set name}/${Embeded Particle}_${fSet}_${RequestID}/
/star/embed/embedding/${Trigger set name}/${Embeded Particle}_${fSet}_${RequestID}/
/star/data18/embedding/${Trigger set name}/${Embeded Particle}_${fSet}_${RequestID}/
3) If they exist, you can further check whether all the ${fSet} from 'fSet min' to 'fSet max' are there. If all exist,
you can start to use it. If none of them are there or only some fraction of 'fSet' are there, write to the embedding list
and ask the Emebedding Coordinator to restage it to there from HPSS.
4) If you can not find the embedding data in this directory, the data must be in HPSS. Unfortunately, there is not a full list of data stored in HPSS yet. (For some data produced at RCF, Lidia maintains the list of embedding samples produced at RCF.) Please write to the embedding list, provide the request ID, Particle Name, file type (minimc.root, MuDst.root or event/geant.root), and ask the Embedding Coordinator to restage the data to NFS disk for you.
Operations (OBSOLETE)
The full list of STAR embedding requests (Since August, 2010):
http://drupal.star.bnl.gov/STAR/starsimrequest
The operation status of each request can be viewed in its page and history.
The information below (and in sub-pages) are only valid to OLD STAR embedding requests.
Request status
- Currently pending requests
- Old Requests summary (circa Nov 2006)
New requests
Current Requests
Current Requests were either submitted using the cgi web interface before it had to be removed (September 2007) or via email to the embedding-hn hypernews list.
As such there is not a single source of information. The excel spreadsheet here is a summary of the known requests as of August 2008 (also pdf printout for those without access to excel). The co-ordinator should keep this updated.
Heavy flavour have also kept an up to date page with their extensive list of requests. See here.
[The spreadsheet was part of a presentation (ppt|pdf) in August 2008 on the state of embedding at the end of my tenure as EC - Lee Barnby]
New Requests
In the near future we hope to have a dedicated drupal module available for entering embedding (and simulation) requests. This will have fields for all the required information and a work flow which will indicate the progress of the request. At the time of writing (August 2008) this is not available. A workable interim solution is to make requests using this page by selecting the 'Add Child Page' link. One should then write a page with the information for the request. Follow-up by members of the embedding team and requestors canthen use the 'Comment' facility on that page.
The following is required
- pt range
- pt distribution (flat, exponential)
- rapidity
- ...
- (to be completed)
Old Requests
Overview
This page is intended to provide details of embedding jobs currently in production.
| ID | Date | Description | Status | Events | Notes |
| 1121704015 | Mon Jul 18 12:26:55 2005 | J/Psi Au+Au | Open | pgw Heavy | |
| 1127679226 | Fri Sep 16 20:34:17 2005 | Photon embedding for 62 GeV AuAu data for conversion analysis | Open | pgw High Pt | |
| 1126917257 | Sun Sep 25 16:13:46 2005 | AMPT full chain | Open | pgw EbyE | |
| 1130984157 | Wed Nov 2 21:15:57 2005 | pizero embedding in d+Au for EMCAL | Open | pgw HighPt | |
| 1138743134 | Tue Jan 31 16:32:14 2006 | dE/dx RUN4 @ high pT | Done | pgw Spectra | |
| 1139866250 | Mon Feb 13 16:30:50 2006 | Muon embedding RUN4 | Test Sample | pgw Spectra | |
| 1139868572 | Mon Feb 13 17:09:32 2006 | pi+/-, kaon+/-, proton/pbar embedding to RUN5 Cu+Cu 62 GeV data | open | pgw Spectra | |
| 1144865002 | Wed Apr 12 14:03:22 2006 | Lambda embedding for spectra (proton feeddown) | Open | pgw Strangeness | |
| 1146151888 | Thu Apr 27 11:31:28 2006 | pi,K,p 200 GeV Cu+Cu | Open | pgw Spectra | |
| 1146152319 | Thu Apr 27 11:38:39 2006 | K* for 62 GeV Cu+Cu | Open | pgw Spectra | |
| 1146673520 | Wed May 3 12:25:20 2006 | K* for 200 GeV Cu+Cu | Done | pgw Spectra | |
| 1148574792 | Thu May 25 12:33:12 2006 | Anti-alpha in 200 GeV AuAu | Closed | pgw Spectra | |
| 1148586109 | Thu May 25 15:41:49 2006 | He3 in 200GeV AuAu | Test Sample | pgw Spectra | |
| 1148586313 | Thu May 25 15:45:13 2006 | Deuteron in 200GeV AuAu | Done | pgw Spectra | |
| 1154003633 | Thu Jul 27 08:33:53 2006 | J/Psi embedding for pp2006 | Open | pgw Heavy | |
| 1154003721 | Thu Jul 27 08:35:21 2006 | Upsilon embedding for pp2006 | Open | pgw Heavy | |
| 1154003879 | Thu Jul 27 08:37:59 2006 | electron embedding for Cu+Cu 2005 | Test Sample | pgw Heavy | |
| 1154003931 | Thu Jul 27 08:38:51 2006 | pi0 embedding for Cu+Cu 2005 for heavy flavor group | open | pgw Heavy | |
| 1154003958 | Thu Jul 27 08:39:18 2006 | gamma embedding for Cu+Cu 2005 for heavy flavor group | open | pgw Heavy | |
| 1154004033 | Thu Jul 27 08:40:33 2006 | electron embedding for p+p 2005 for heavy flavor group (e-h correlations) | Test Sample | pgw Heavy | |
| 1154004074 | Thu Jul 27 08:41:14 2006 | pi0 embedding for p+p 2005 for heavy flavor group (e-h correlations) | Test Sample | pgw Heavy | |
| 1154626301 | Thu Aug 3 13:31:41 2006 | AntiXi Cu+Cu (P06ib) | Done | pgw Strangeness | |
| 1154626418 | Thu Aug 3 13:33:38 2006 | Xi Au+Au (P05ic) | Done | pgw Strangeness | |
| 1154626430 | Thu Aug 3 13:33:50 2006 | Omega Au+Au (P05ic) | Done | pgw Strangeness | |
| 1156254135 | Tue Aug 22 09:42:15 2006 | Phi in pp for spin-alignment | open | pgw Spectra | |
| 1163565625 | Tue Nov 14 23:40:25 2006 | muon CuCu 200 GeV | open | pgw Spectra | |
| 1163627909 | Wed Nov 15 16:58:29 2006 | muon CuCu 62 GeV | open | pgw Spectra | |
| 1163628205 | Wed Nov 15 17:03:25 2006 | phi CuCu 200 GeV | Test Sample | pgw Spectra | |
| 1163628539 | Wed Nov 15 17:08:59 2006 | K* pp 200 GeV (year 2005) | Open | pgw Spectra | |
| 1163628764 | Wed Nov 15 17:12:44 2006 | phi pp 200 GeV (year 2005) | Open | pgw Spectra |
runs for request 1154003879
min-bias run list from Anders.6031103 6031104 6031105 6031106 6031113 6032001 6032003 6032004 6032005 6032011 6034006 6034007 6034008 6034009 6034014 6034015 6034016 6034017 6034108 6035005 6035006 6035007 6035009 6035010 6035011 6035012 6035013 6035014 6035015 6035016 6035026 6035027 6035028 6035030 6035032 6035036 6035108 6035111 6036012 6036014 6036016 6036017 6036019 6036020 6036021 6036022 6036024 6036025 6036028 6036036 6036039 6036043 6036044 6036045 6036098 6036102 6036103 6036104 6036105 6037009 6037010 6037013 6037014 6037015 6037016 6037017 6037018 6037019 6037025 6037026 6037028 6037029 6037030 6037031 6037033 6037039 6037040 6037046 6037047 6037048 6037049 6037050 6037053 6037054 6037071 6037073 6037075 6037077 6038085 6038088 6039033 6039034 6039040 6039132 6039134 6039135 6039136 6039138 6039139 6039141 6039142 6039143 6039144 6039145 6039154 6040001 6040003 6040004 6040007 6040008 6040009 6040026 6040043 6040044 6040045 6040048 6040050 6040051 6040052 6040054 6040055 6040056 6040057 6040058 6041014 6041015 6041020 6041021 6041022 6041023 6041025 6041027 6041028 6041031 6041034 6041035 6041036 6041062 6041063 6041064 6041065 6041066 6041091 6041092 6041093 6041097 6041099 6041102 6041103 6041105 6041118 6041120 6042003 6042006 6042008 6042009 6042010 6042012 6042014 6042015 6042016 6042018 6042024 6042046 6042047 6042048 6042049 6042052 6042054 6042055 6042057 6042059 6042060 6042101 6042105 6042109 6042110 6042111 6042112 6042113 6043010 6043011 6043014 6043017 6043019 6043020 6043021 6043022 6043026 6043027 6043039 6043043 6043045 6043054 6043057 6043058 6043060 6043063 6043080 6043082 6043085 6043090 6043094 6043095 6043097 6043098 6043100 6043101 6043111 6043112 6043113 6043114 6043115 6043117 6043118 6043119 6043120 6043121 6043122 6043123 6044001 6044010 6044011 6044012 6044015 6044016 6044020 6044024 6044025 6044026 6044027 6044028 6044044 6044045 6044046 6044047 6044049 6044050 6044053 6044054 6044055 6044058 6044059 6044075 6044079 6044081 6044083 6044084 6044085 6044086 6044088 6044089 6044090 6045008 6045026 6045027 6045029 6045033 6045042 6045070 6045072 6045075 6045076 6045077 6045078 6045079 6045080 6045081 6045082 6046004 6046005 6046014 6046016 6046017 6046018 6046022 6046028 6046035 6046037 6046038 6047017 6047020 6047022 6047025 6047026 6047037 6047039 6047040 6047044
Submit a new embedding request
Before submitting a new request, a double-check in the simulation request page is recommended, to see whether there are existing requests/data can be used. If not exist, one need to submit a new request. Please first read the 'Appendix A' in the embedding structure page for the rules to follow.
If the details of the embedding request has been thoroughly discussed within the PWG and hence approved by the PWG conveners. Please the PWG convener add a new request in the simulation request page and input all of the details there.
There are some key information that must be provided for each embedding request. (If you can not find some of the following items in the form, simply input it in the 'Simulation request description' box.)
Detailed information of the real data sample (to be embedded into).
- Trigger sets name. for example, "AuAu_200_production_mid_2014", a full list of STAR real data trigger sets can be found in the data summary page.
- file type. for example, "st_physics", "st_ht", "st_hlt", "st_mtd", "st_ssdmb". Please note that ONLY those "st_*_adc" files can be used for embedding production. Due to limitation of disk space, we usually sample 100K events (~200-500 daq files, depending on trigger and other event cuts), and this set of daq files will be embedded multiple times to reach the desired full statistics (say 1M).
- Production tag. for example, "P15ic"
- Run range with list of bad runs, or a list of good runs. for example, "15076101-15167014"
- trigger IDs of the events to be embedded. for example, HT1 "450201, 450211"
-
vertex cut, and vertex selection method. for example, "|Vertex_z|<30cm", "Vr<2cm", "vertex is constrained by VPD vertex, |Vz-Vz_{VPD}|<3cm", "PicoVtxMode:PicoVtxVpdOrDefault, TpcVpdVzDiffCut:6" or "default highest ranked TPC vertex".
- Other possible event cuts, for example, cut on refmult or grefmult, "refmult>250"
- Each request can only have ONE type of dataset described above.
Details for simulation and reconstruction.
- Particle type and decay mode (for unstable particle). for example, "Jpsi to di-muon"
- pT range of input particle and the distribution. for example, "0-20GeV/c, flat", or "0-20GeV/c exponential"
- pseudo rapidity or rapidity range (the distribution is flat), for example, "pseudo-rapidity eta, -1 to 1", or "rapidity, y, -1 to 1". Please do make clear it is "PSEUDO" rapidly or rapidity, as they are quite different quantities.
- Number of MC particles per event. Usually "5% of refmult or grefmult" is recommended, one can also assign a fixed number, for example "5 particle per event" for pp collisions.
- For event generator embedding request, like Pythia/HIJING/StarLight in zero-bias events, please provide the generator version at least. For example, "Pythia 8.1.62". The PWG embedding helper or PA's are fully responsible to tune-up the parameters for the generator in such embedding requests.
- One can add special requirement for production chain in 'BFC tags' box, for example turn off IFT tracking in Sti.
- Please indicate whether EMC or BTOF simulator is needed.
Finally, please think carefully about the number of events! The computing resources (i.e. the CPU cores and storage) are limited !
It is acceptable to modify the details of the request afterwards, although it will be of great help if all above detailed information can be provided when a new request is submitted, in order to avoid the time waste in communications. If this is inevitable, please notify the Embedding Coordinator immediately if the details of a request is modified, especially when the request is opened.
Weekly embedding meeting (Tuesday 9am BNL time)
We start weekly embedding meeting at Tuesday 9am US eastern time, to discuss all embedding related topics.
The Zoom link can be found in below:
Topic: STAR weekly embedding meeting
Time: Tuesday 09:00 AM Eastern Time (US and Canada)
Join ZoomGov Meeting
https://bnl.zoomgov.com/j/1606384145?pwd=cFZrSGtqVXZ2a3ZNQkd1WTQvU1o0UT09
Meeting ID: 160 638 4145
Passcode: 597036
Meetings in 2025:
- Meeting August 5, 2025
- Recording: https://bnl.zoomgov.com/rec/share/-aWyB-_If-B80-96qqcJkCp5elfCWmw-E8Ekd63Yz_VtEXYkkiXDAVBxsehURc0Z.lIyEBwhFTIpPK4tK
Passcode: YFCH3?jQ
Agenda:1) Status and planning of embedding production
- Meeting July 29, 2025
- Recording: https://bnl.zoomgov.com/rec/share/-tDAfIhVojy8L4GaKlG7NyTYoElqNaz4NfDhV3E7wMUilA1l1Rbu4GKRKFO8l6Gy.RnJ7SINZVQoa12ys
Passcode: SAy%JE&9
Agenda:1) Status and planning of embedding production
- Meeting July 22, 2025
- Recording: https://bnl.zoomgov.com/rec/share/sBuYZb6hYCUPy-IBLwzCI_0I4wiHbQvWNf1JsRRDijigJWsxEDNWl4WngdCEjx-y.GLRwgMTyfAgZ_f2_
Passcode: ZLs4F$?@
Agenda:1) Status and planning of embedding production
- Meeting July 15, 2025
- Recording: https://bnl.zoomgov.com/rec/share/NcxJ1wwrbc554XwQ_kQi9_oVC1VTrYxKQ_DOJ3ML3i6FUaL8ZEOY4BlSE-YVwslT.F2FGfhDogr24Ln-N
Passcode: SB=d7%36
Agenda:1) Status and planning of embedding production
- Meeting July 8, 2025
- Recording: https://bnl.zoomgov.com/rec/share/2EHOfztLozxKmIZdSmqUJlAIJrfG-vdeK6-BVQz2sga5R6Z3MijSjJAypfI2o8oO.UQGezbn_eJXLAAtO
Passcode: #2.i@qL8
Agenda:1) Status and planning of embedding production
- Meeting July 1, 2025
- Recording: https://bnl.zoomgov.com/rec/share/CljeYpUO3ID_6TNaU05vDlz20fBv8AlOhLABndr6yPvVEe3NoyAE1FoDeFt-5QsC.N5PK4vRDKbha_7Pz
Passcode: hkd+tPP4
Agenda:1) Status and planning of embedding production
- Meeting June 24, 2025
- Recording: https://bnl.zoomgov.com/rec/share/VHv3sGgEJ2kM4Mdh4N4WsGBZVvAiS4J_j7Y3tmx9WQwdAcHPbTOeiu_d56bRnE5B.sNmvNpva4LMSwN1J
Passcode: j#^7gcAJ
Agenda:1) Status and planning of embedding production
- Meeting June 10, 2025
- Recording: https://bnl.zoomgov.com/rec/share/OkTnUWdIBlModNU8W2-AjaYQb0QSL1iNufi7QygL9f1xRJRGXAmsUdKtp5dWu8wS.T8PERgku_C206Ctc
Passcode: qCy0h^@u
Agenda:1) Status and planning of embedding production 2) backup of embedding code
- Meeting June 3, 2025
- Recording: https://bnl.zoomgov.com/rec/share/T0ZKVYly0GufhVBUDo_ceVUC8x98bB3N-dyBGiyg9LXjm7iLgptp8NnVwRZTBrV7.dG4Znnwu3ibjSR3i
- Meeting May 27, 2025
- Recording: https://bnl.zoomgov.com/rec/share/X89ira4U_4D_ldMnIqGYJ_Frr-grn9Ytyqb2-PEOMVxPkaXFKGRzCzBDonNaiFSp.D3OH10WNq-ezcCNk
Passcode: vF7Db=*G
Agenda:1) Status and planning of embedding production
- Meeting May 20, 2025
- Recording: https://bnl.zoomgov.com/rec/share/CqRPyJqN5f__IxVgDsiRgzWE6XOrOWukvf6yDCztG9rhdfgVrmHv0c9jSQ53QLWw.6Pau3F0OZ6ol5x6c
Passcode: WiZ8T6+A
Agenda:1) Status and planning of embedding production
- Meeting May 13, 2025
- Recording: https://bnl.zoomgov.com/rec/share/kxNjgpfsZX02aNZfSNmawdQn7StSUQS28WzEQCOu8JTQ-i3mh1HnChPj3HKJ0NFj.A_stYyHsb6RQO_6J
Passcode: ?E1r^gcY
Agenda:1) Status and planning of embedding production
- Meeting May 6, 2025
- Recording: https://bnl.zoomgov.com/rec/share/Eh-d76u6fvvc9RqDs8UhmSC-mMRdWZZfaJuOMpPp8jdKsXN_B8Ei9upDrspFuEHY.3Nv-jVVxH84toZXQ
Passcode: wGa$bP4C
Agenda:1) Status and planning of embedding production
- Meeting April 29, 2025
- Recording: https://bnl.zoomgov.com/rec/share/3SeE0vahaP6AzvSG4w0jTqW3KuDbUpbMkPDb4VZTBlTQMDNGtrsJIMOh6a_ue4gX.O4tjR0req1rRn1v-
Passcode: hcgQ@cj5
Agenda:1) Status and planning of embedding production
- Meeting April 22, 2025
- Recording: https://bnl.zoomgov.com/rec/share/mdwYz9jmA1wjKQGp9BY-pT0twy9QNyqF9PaISHdIY8542IjQrj1f1CdMOKFL_GHQ.qfW_-YE9uFCD6Jmi
Passcode: dyx%2l%5
Agenda:1) Status and planning of embedding production
- Meeting April 15, 2025
- Recording: https://bnl.zoomgov.com/rec/share/31XJuN_8zhHR5h4cD0cp0kpbpC5R-gWpc2wN3OE_cQLk6ikFz6ND_sq14NzcwUWV.INtG5fo0P0u-T3vw
Passcode: +Z070A0+
Agenda:1) Status and planning of embedding production
- Meeting March 25, 2025
- Recording: https://bnl.zoomgov.com/rec/share/QTh7-_Y_q4Oh2aF4l8MympZ8GHC0d_Qr57yI4v_0JqX9iJu4JZzjeZVaL2ViEbrE.J0vHtoP4FUt6U4Fs
Passcode: @1EMYn^f
Agenda:1) Status and planning of embedding production
- Meeting March 18, 2025
- Recording: https://bnl.zoomgov.com/rec/share/ziUz9DPbOdwJWjcwmQgDucQ5twkfEaOBL-W_rmspIRM8ArGb7XMN87GKfTvs1Pw6.4b617t8gKa1hF730
Passcode: #*36PhjP
Agenda:1) Status and planning of embedding production
- Meeting March 11, 2025
- Recording: https://bnl.zoomgov.com/rec/share/fGMj6M_z1x8eQ832GszpT21DfRJ2ayianolJTIxMH50havXXSAwRp_FuGSXYGh08.XXQAsRfj1DvaQ48u
Passcode: z6TkP6x#
Agenda:1) Status and planning of embedding production
- Meeting February 25, 2025
- Recording: https://bnl.zoomgov.com/rec/share/0qGYCtYMSLtwhitEeGbGNmnU61HsxJHVJ9vmGCZcBNyQjt9sdebck-KJa6RngdbA.xQ2VPOlXf-xj_Fvr
Passcode: &L7#Rj4u
Agenda:1) Status and planning of embedding production
- Meeting February 18, 2025
- Recording: https://bnl.zoomgov.com/rec/share/FOBOU8TKE58iO3w_gUtBNQL-z_rPQ-j0IcEgj5ptdw72-I2Dd2Bu9I74c5NePoJU.U-z-zrZSPf3SPkZV
Passcode: g$*?022M
Agenda:1) Status and planning of embedding production
- Meeting February 11, 2025
- Recording: https://bnl.zoomgov.com/rec/share/QvHgqXvnllg5mrKSHPmqfnC7dMLb902QPA27eZ-2x_bJ86RAypGA0Iirz7F7WlnL.YZTrKGZ4dQLvMjjk
Passcode: K2Dr%y8^
Agenda:1) Status and planning of embedding production
- Meeting February 4, 2025
- Recording: https://bnl.zoomgov.com/rec/share/aqeMOuMzbAqxt2D13CXYStZpfZGBp7CeNOawLtXfj47zhBPMCxKxQqz0U_Qz8LbS.qeO-jYGhSgN_NmkL
Passcode: @8#A=1t$
Agenda:1) Status and planning of embedding production
- Meeting January 21, 2025
- Recording: https://bnl.zoomgov.com/rec/share/_sJdfvoByiHN25LOXfsIIGXApvfKP6oBJy-z_DB-RvmD5-zRTjuYCDZ0-6zFtymq.bYB2TAPEpL159QTD
Passcode: P#F0jc&z
Agenda:1) Status and planning of embedding production
- Meeting January 14, 2025
- Recording: https://bnl.zoomgov.com/rec/share/aG8bR7cMOccEdYIV64el6WnsG5qqAkBKIJVPThH4Bads5gwXSlTwKW38mWKaLt9A.dcK1s2-ueypS8KIs
Passcode: GL^CHK2*
Agenda:1) Status and planning of embedding production
-
Meeting January 7, 2025
Recording: https://bnl.zoomgov.com/rec/share/p3ZK8040XYYtIKCmscD-zzkOMKXdoAkm3NKH9XCziJIZQX8knabRg9CAKlHx863N.7eQT8CgiNxFjIYwq
Passcode: w3Na*4r3
Agenda:1) Status and planning of embedding production
Meetings in 2024:
- Meeting December 17, 2024
- Recording: https://bnl.zoomgov.com/rec/share/M98X-5Arz-AvV4d9UI1oLLsd_MC2XZQYgyRgfvf5G6DZJHBwMyb0dfJu4imqZFpG.p7OA7lYXRJuupGk2
Passcode: 7=hsgCj7
Agenda:1) Status and planning of embedding production
- Meeting December 10, 2024
- Recording: https://bnl.zoomgov.com/rec/share/OhK2auDMNWMvJvycQn2DvMGOZAsLnLoUL6pcaXcV6x1p6TtK2ScHy0T8KodmlkhX.0qYUzQD0ldBk1ujc
Passcode: B*J1c.N8
Agenda:1) Status and planning of embedding production
- Meeting November 26, 2024
- Recording: https://bnl.zoomgov.com/rec/share/5pP_HlklFPhQrhTeCpXYMncAeqAEEhs0pthuI6znBfSd5jHZJaOQc4NGb-T16VvW.m1lSoSVOGJEWKwbX
Passcode: MB4yxJ*y
Agenda:1) Status and planning of embedding production
- Meeting November 19, 2024
- Recording: https://bnl.zoomgov.com/rec/share/L9ca_hh5qv_RKBH3RVWzPTfcqDMv3dxmKebdFPWZ5WnHcrbc5sEFFadl-n0552Qr.yRnSmmqUGQV_82H_
Passcode: hYA2MW=E
Agenda:1) Status and planning of embedding production
- Meeting November 12, 2024
- Recording: https://bnl.zoomgov.com/rec/share/00e2PVyaFb45TvARixMNIwnlkHaTnz-V3L4qf34JQUpBG64dvj-F6KjzsG7eELH-.jZjL6Zd_VwXWuhJl
Passcode: hL+B?#5@
Agenda:1) Status and planning of embedding production
- Meeting November 5, 2024
- Recording:https://bnl.zoomgov.com/rec/share/X8IOOJE4Dd5W7dJbNqmGiRHNGWHOUYAPi8Gr93nE_36gQivE75uHZZZoEOP9rGGa.Wz0D4QFha72hj3pk
Passcode: i*g7YY3C
Agenda:1) Status and planning of embedding production
- Meeting October 29, 2024
- Recording: https://bnl.zoomgov.com/rec/share/GSh5VqhJ0utOoLaWXwlhUslHbuNg_U5NfziAAF055C4f8CuBgBhfJL4E8o5OWRA-.ONnicKQwiWoz327e
Passcode: h6gZ^Znt
Agenda:1) Status and planning of embedding production
- Meeting October 15, 2024
- Recording: https://bnl.zoomgov.com/rec/share/uAIOTRfQlT5CRCsQHVT8tImDIvZAxR-XZoSSSTyHKjnIihT6fjhP0S20sWRJ3NAH.ghVv14EicAmibalL
Passcode: %F*9B1#.
Agenda:1) Status and planning of embedding production
- Meeting October 8, 2024
- Recording: https://bnl.zoomgov.com/rec/share/WwjLgK3B41p2HFisXC5Ig0nlNkQcj5SOR8aD70FtdU8yzKSjQ5KTzS78wq3jID0g.jgV3n2yYR2sPkvSI
Passcode: RDNr&V%0
Agenda:1) Status and planning of embedding production
- Meeting September 24, 2024
- Recording: https://bnl.zoomgov.com/rec/share/xDche1El2zE57Qorl6ZkkjJDZtInzm8tM05S-w1awkQ62v0TRB5jrBmkRn_orhe9.N9Kjki195-JLtvHS
Passcode: #QeY!0Ld
Agenda:1) Status and planning of embedding production
- Meeting September 3, 2024
- Recording: https://bnl.zoomgov.com/rec/share/ybXe4plUB3AQOX8hZ3c0M6GP_dhq5v_tj5mOo4GNxLfZ0vIvgqInyizmsWhMW1f6.fz1mvYENvEneSZ0c
Passcode: 0SN6F&E@
Agenda:1) Status and planning of embedding production
- Meeting August 27, 2024
- Recording: https://bnl.zoomgov.com/rec/share/0r7TcUFM8zkkTwStohOn6W-5s0pLvgG6eZ-8y0Tzk3apnXrE_AFeUJ8ZR9Qk9tj0.CPdmSMpvIfzmU1Ov
Passcode: 5X?!Rhmj
Agenda:1) Status and planning of embedding production
- Meeting August 20, 2024
- Recording: https://bnl.zoomgov.com/rec/share/qcsPIH987hJ_imLT3pnUxd2G3ewxRru0PLyIjHe8RWMUH6EVFgvnpEYjcKTO688H.ZLmwAUPTZaOue5qZ
Passcode: ^9bMy0Fs
-
Agenda:1) Status and planning of embedding production
-
- Meeting August 13, 2024
- Recording: https://bnl.zoomgov.com/rec/share/NjiD0Aj6MNx0PB6Fes_rB6lHpoObCD_gvLYkSlfbdpQ2px_Vdoe42jp_g8-v5o9n.AGpGQlo0ZAIZ_rqP
Passcode: tpV4ww^9
Agenda:1) Status and planning of embedding production
- Meeting August 6, 2024
- Recording: https://bnl.zoomgov.com/rec/share/Tio6ajxy1IaSYTw4l6-H5RiqYAASXg0OBHcpClAgyH-MwqTcTAmg17MP4cMfr296.8s9Sjr89YoNlZUuD
Passcode: C*K#.$2U
Agenda:1) Status and planning of embedding production
- Meeting July 30, 2024
- Recording: https://bnl.zoomgov.com/rec/share/mPpvfKqwmcvf_N3HmSXIDoGYnvd5lnlIxGBfBwJiKREeNXmCvkKZmAcsyFT3p6W5.KesWegECSFK_Q5_z
Passcode: P=.kN*94 (status around 15min~20minute)
Agenda:1) Status and planning of embedding production
- Meeting July 23, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/BQNzkefqPku0_rVX6ImcmjuBOOCYNgNZ6vF5qIWPna5wEd9Q8G-vQ8HLx5fbkbFg.mk9Cwmhw5CxZn0pt
Passcode: y@2H0^xV
Agenda:1) Status and planning of embedding production
- Meeting July 16, 2024
- Recording: https://bnl.zoomgov.com/rec/share/RTP-QJz0cfiZfP9QQYKF7c_LgIaGB8nxOi8eFhwvG-p4M-NE5siPBYnHgyu9j_iR.ixFKLYAwERfRUnuO
Passcode: t9aA.3VJ
Agenda:1) Status and planning of embedding production
- Meeting July 9, 2024
- Recording: https://bnl.zoomgov.com/rec/share/Gi92JOeJ8JRM4fLC1fbyMlLMtsXJsbBVqfK5gLmsBw1IA1ezs8HJIVxTxgrgpYI5.CQ95RoEbhnHNViuX
Passcode: Li+85D7a
- Agenda:1) Status and planning of embedding production
- Meeting July 2, 2024
- Recording:https://bnl.zoomgov.com/rec/share/pFFAwg3fEhH6B-65lKqrWtUS_6RAsTBfticZWYIkN0Kryy3DGW-CtAzZnJ04vEjj.OKvddins359m7tws
Passcode: K+#3E3Kv
Agenda:1) Status and planning of embedding production
- Meeting June 25, 2024
- Recording: https://bnl.zoomgov.com/rec/share/5JEPs-KwYWUK3RJrJpIk7031gDTJmfcpIsYP1624s3S6DLtEAbqKlivsOkOSzdv3.JEjIReugeExEAAKs
Passcode: S*.Zy0qW
Agenda:1) Status and planning of embedding production
- Meeting June 18, 2024
- Recording: https://bnl.zoomgov.com/rec/share/HYjG3OKAXKzskh0unTdDdAkQIVtAX1K5ULLjQgW07jniMzZuK8ieMsRoutinjSPg.fKOISCSK2MxEuGPk
Passcode: kD=&m2E!
Agenda:1) Status and planning of embedding production
- Meeting June 11, 2024
- Recording: https://bnl.zoomgov.com/rec/share/8W25-Nm-440wOfh54Cn7uBvEJoZLU00P3MqFbRzhQor3ymNuEPO2hqy1BfZldasi.PDziwtupxHS2Aaoi
Passcode: @V*Ho39G
Agenda:1) Status and planning of embedding production -
- Meeting June 4, 2024
- Recording: https://bnl.zoomgov.com/rec/share/D5cKyf5biJ6qGsy1WTHw_HN4tG_nxk9X8-iIGQOOu_U1r9x-0a3I2sC6szonZDe-.ElsZbETCKGZO_DII
Passcode: 2!bipSh&
Agenda:1) Status and planning of embedding production
- Meeting May 28, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/5OFU1BUjipIcqTAzE1btYytredl3MIxeiifxP9yytBOv3RFhtpHi9cSDCZ6ObitD.wrlRhYIzFRDoCdtg
Passcode: ^Z5u*E.h
Agenda:1) Status and planning of embedding production -Xionghong Maowu, Xianglei
2) QA plot and discussion on e+e- embedding at 9.2 GeV, Zhen et al
3) Discussion on run12 jet substructure embedding set up, -Isaac, Youqi et al
- Meeting May 21, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/6_NiaPCD7-FcfusDiuR2KahRGwbeirpoJQPxM9nAk1tCit5YowzPxdFNx1MEXo2a.aKeBuIYm1wfS15Zl
Passcode: nF%2^bxU
Agenda:1) Status and planning of embedding production - Xianglei, Pavel, Xionghong, Maowu - Meeting May 14, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/VZlIAA3uxx8xjALEYr4Od-rvfUGFLxZ0d3pr4sMqwX_De0gaYbmRmfsx96cAQXhd.PATlmUeE8B48srn0
Passcode: F!wh0DTF
Agenda:1) Status and planning of embedding production - Xianglei
- Meeting May 7, 2024
-
Passcode: 9K*C!?KZ
Agenda:1) Status and planning of embedding production - Xianglei
- Meeting Apr. 30, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/J2KbzQC5u1hDgVeM4CIGadVJCc7zBOKjVdX2fW8n-sOqK9R3XJZIHDfRTtRozL0_.uC0ZIS5Qa28NX6Ke
Passcode: KP*K5vNE
Agenda:1) Status and planning of embedding production - Xianglei
- Meeting Apr. 23, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/c_yJJlq6LJez3pRRCHiQQqEZd_h3XetZAC9d4lSdzF2ac4Tpe04f4GJ09xwM_0il.CoPzy7pcVg4eN36n
Passcode: EAKp1*z%
1) Welcome Pavel Kisel to join the embedding team as deputy -Qinghua
2) Status and planning of embedding production - Xianglei
- Meeting Apr. 16, 2024
- Recording: https://bnl.zoomgov.com/rec/share/4cJxLv9Zdd1f24sjSHL_rL6CDScvXpQz8WfaaqcDETjOx1WdWl4-IbCta3KRSRqX.5QgqOO4bfQL7xg0t
Passcode: ^$d54Gqm
1) Status of embedding production - Xianglei
2) Discussion of embedding planning -all
- Meeting Apr. 9, 2024
-
Passcode: bBQ=8@cS
1) Status of embedding production - Xianglei
- Meeting Apr. 2, 2024
- Recording: https://bnl.zoomgov.com/rec/share/2sS3NxMOPp2pIXxEqKIzmouSL56lwzq1s3n1OHKqKI7OpMrNKnJO8YGAX9LY638o._EWbhxENm5lsovia
Passcode: WwLTH@@7
1) Status of embedding production - Xianglei
2) Discussion of embedding plan for coming conferences -all
- Meeting Mar. 26, 2024
- Recording: https://bnl.zoomgov.com/rec/share/cCdNqJJPP_R5IZSUcjh3qAFRv8SdKzMIf69qyxWQM8kE7G8ymSplc33NgsVkyeGS.T8NgCp1bw8bmxij6
Passcode: +Z&Yn&A2
Agenda: 1) Status and planning of embedding production - Xianglei
- Meeting Mar. 12, 2024
- Recording: https://bnl.zoomgov.com/rec/share/GqXYxb8WnlbC8uxoqyxNcVfzzW2QllEx-NxK20o1CFQdqIf5gvPNvjYgPL5xzqtk.OLWojmdiKAjSU5WS
Passcode: 5c+D0fVa
Agenda: 1) Status and planning of embedding production - Xianglei et al
- Meeting Mar. 5, 2024
- Recording: https://bnl.zoomgov.com/rec/share/Auq1KjlVZmMZ-qaf68mBH70cKP897wEEAJABv9cDEQe69VpFmncxLJPAmxdDrceS.Q7CLuQ0hJwDMRKdn
Passcode: %&d2nRb$
Agenda: 1) Status of embedding production-Xianglei; 2) Remaining embedding request & priority list- Sooraj and all
- Meeting Feb. 27, 2024
- Recording: https://bnl.zoomgov.com/rec/share/m77wid3nSeS4aTODff4MWZYrrRCEOaBSJhy0stnKMZ7uuZ7DmN5cTjlZGI7wpTTZ.dp0d0vWa9V8kULo2
Passcode: %rGQ8w5%
Agenda: 1) Status and planning of embedding production - Xianglei
2) an example of base QA for embedding helpers -Yi Fang
- Meeting Feb. 20, 2024
- Recording: https://bnl.zoomgov.com/rec/share/gHSyg2oO-23SZqpyd52Bp03YRJq4g8IhLmJUakqbrcA5rbSPOMAIza8zhf0xdVKH.L5n4hvWXE373wWcY
Passcode: R7U@0z*D
1) Status of embedding production - Xianglei, all
2) Planning of remaining requests - all
- Meeting Jan. 30, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/WEyCkpky3OTH0N-lE6ggswGTT0ydeJwMN2UXFSRSMqK7y8W5s4sKqiwYz5P3yS0R.kOJlJx5OXGG5Mzh4
Passcode: 50+kgTW*
Agenda: 1) Status of embedding (17.3 GeV production completed. ) Xianglei
2) embedding production planning
- Meeting Jan. 23, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/Zjnp8Gp2Nwk-eNH13UoxDnq88Kela_JOg_V6QMGjEZZGjqsM5ZokGtx80lNHIspQ.pIIXa6YvN0wgsyx-
-
Passcode: y3?P.tPW
- Agenda: 1) Status and planning of embedding production (17.3 GeV tuning finished, testing sample available in 1~2 days) -Xianglei
- Meeting Jan. 9, 2024
- Recordings: https://bnl.zoomgov.com/rec/share/sM2zgy4Ke3rIWK3bpSVUVz77bhu_OW0sXd_XvI4PmU3ZwgeBM0TNMEm8Jcv_6Gii.pGH5mTtWkGiwd4q9
Passcode: 9EF4Y$ei
Agenda: 1) Status of embedding (found a DB tracking issue from Dec. 14, temporary patch applied. 17.3 GeV tuning underway. )
2) embedding production planning
*********************************Meeting in 2023***************************
- Meeting Dec. 19, 2023
-
Recording: https://bnl.zoomgov.com/rec/share/PXEyMAM1wTqIoYNEQO__Ex9X0Zv2YVZfVIO-vFu-RtwFiLmoeXDAYO4yzDJgmFX_.ozgESVwcNJmoONsQ
Passcode: vcq*?*60
- Meeting Dec. 12, 2023
-
Recording: https://bnl.zoomgov.com/rec/share/LAhvI5YxeVqNmlvoU_F_kCwX9J_FKo0g20BAiK3R8mvv7nV30uMzdbDZ5gLYzBVY.F2I4pteWl6On0yRu
Passcode: Sbi46+=kAgenda: 1) Status of embedding production - Xianglei (parameters tuned and testing sample for 11.5 GeV produced)
2) Discussion on the priority list of embedding request - Sooraj, Xianglei et al
- Meeting Dec. 05, 2023
-
Passcode: @3Q1yze!
1) Embedding status and feedback- Xianglei (9.2GeV sample produced, 11.5 GeV starts retuning )
2)Reproduction request of run 12 pp 200 GeV embedding -Youqi
-
Meeting Nov. 28, 2023,
Recording:https://bnl.zoomgov.com/rec/share/DUfRTTgmVNmFbZsJ6M5RMoYvpTQcf7gq_obTy9vpysPd8KY2GIltJ7FVKe05I46k.V9NNoP7AzwWSY7CG
Passcode: 90j9!kJ6Agenda:
1) Status of embedding production -Xianglei
-summary: Testing sample available for 9.2GeV (pass base QA), need analyzer to look at them before official production. Started with 11.5GeV.
- Meeting Nov. 21, 2023:
- Recording:https://bnl.zoomgov.com/rec/share/sHJguAtToNtF203DkZkiIUp8KavRvyAjzknk8uzuHq6W6iCdjaJVVeN5q7YUJdui.GtE3aSJA8zrxXhs-
Passcode: jZP#xm61
Agenda:
1) Status of embedding production at 9.2 GeV Au+Au collision -Xianglei+all
- Brief summary: A few remaining issue-T0 offset tuning, iTPC gain tuning, transverse diffusion. Testing sample will be available in a few more days for QA.
2) Embedding planning, all
- Meeting Nov. 14, 2023:
- Recording: https://bnl.zoomgov.com/rec/share/mIgXhfQfpP1El77ZMb0NH1bjb0BsZkiaxhUDHKTdpck-IzT0lsiAZFUnM3h7fSfA.JY4xp4iLrd91wduQ
Passcode: Gz^g9*J3
Work Planned (OBSOLETE)
Based on:- An initial meeting held during QM06 (Chair: J.Lauret, L. Barnby, O, Baranikova, A. Rose)
- Email exchange and notes from the meeting (From: J. Lauret, Date: 11/20/2006 22:36, Subject: Summary of our embedding meeting)
- Further EMails from L. Barnby in embedding-hn (Date: 1/18/2007 15:12, Thread ID 84)
| ID |
Task
|
% Complete | Duration | Start | Finish | Assigned people |
|---|---|---|---|---|---|---|
| 1 |
General QA consolidation
|
28% | 109 days? | Thu 11/16/06 | Tue 4/17/07 | |
| 2 | ||||||
| 3 |
Documentation
|
25% | 37 days | Mon 12/18/06 | Tue 2/6/07 | |
| 4 |
Port old QA documentation to Drupal, define hierarchy
|
50% | 4 wks | Mon 12/18/06 | Fri 1/12/07 | Cristina Suarez[10%] |
| 5 |
Add general documentation descriptive of the embedding purpose
|
0% | 2 days | Mon 1/15/07 | Tue 1/16/07 | Lee Barnby[10%],Andrew Rose[10%] |
| 6 |
Add documentation as per the embedding procedure, diverse embedding
|
0% | 2 days | Mon 1/15/07 | Tue 1/16/07 | Lee Barnby[10%],Andrew Rose[10%] |
| 7 |
Import PDSF documentation into Drupal
|
0% | 1 wk | Wed 1/17/07 | Tue 1/23/07 | Andrew Rose[10%] |
| 8 |
Review and adjust documentation
|
0% | 1 wk | Wed 1/24/07 | Tue 1/30/07 | Olga Barranikova[10%],Andrew Rose[5%],Lee Barnby[5%] |
| 9 |
Deliver documentation to collaboration for comments
|
0% | 1 wk | Wed 1/31/07 | Tue 2/6/07 | STAR Collaboration[10%] |
| 10 |
Drop all old documentation, adjust link (redirect)
|
0% | 1 day | Wed 1/31/07 | Wed 1/31/07 | Andrew Rose[15%],Jerome Lauret[15%] |
| 11 | ||||||
| 12 |
Line of authority, base conventions
|
84% | 52 days | Thu 11/16/06 | Fri 1/26/07 | |
| 13 |
Meeting with key personnel
|
100% | 1 day | Thu 11/16/06 | Thu 11/16/06 | Jerome Lauret[10%],Olga Barranikova[10%],Andrew Rose[10%],Lee Barnby[10%] |
| 14 |
Define responsibilities and scope of diverse individual in the embedding team
|
100% | 1 mon | Mon 12/4/06 | Fri 12/29/06 | Jerome Lauret[15%],Olga Barranikova[6%] |
| 15 |
Define file name convention, document final proposal
|
50% | 2 wks | Mon 1/15/07 | Fri 1/26/07 | Jerome Lauret[6%],Lee Barnby[6%],Lidia Didenko[6%],Andrew Rose[6%] |
| 16 | ||||||
| 17 |
Collaborative work
|
45% | 60 days | Mon 1/22/07 | Fri 4/13/07 | |
| 18 |
General Cataloguing issues
|
0% | 9 days | Mon 1/29/07 | Thu 2/8/07 | |
| 19 |
Test Catalog registration, adjust as necessary
|
0% | 4 days | Mon 1/29/07 | Thu 2/1/07 | Lidia Didenko[20%],Jerome Lauret[20%] |
| 20 |
Extend Spider/Indexer to include embedding registration
|
0% | 1 wk | Fri 2/2/07 | Thu 2/8/07 | Jerome Lauret[10%] |
| 21 |
Bug tracking, mailing lists and other tools
|
69% | 60 days | Mon 1/22/07 | Fri 4/13/07 | |
| 22 |
Re-enable embedding list, establish focus comunication at PWG level and user level
|
75% | 3 mons | Mon 1/22/07 | Fri 4/13/07 | Jerome Lauret[10%] |
| 23 |
Establish embedding RT system queue
|
0% | 1 day | Tue 1/23/07 | Tue 1/23/07 | Jerome Lauret[5%] |
| 24 |
Exercise embedding RT queue, adjust requirement
|
0% | 4 days | Wed 1/24/07 | Mon 1/29/07 | Andrew Rose[10%] |
| 25 |
Establish data transfer scheme to a BNL disk pool
|
0% | 22 days | Mon 1/22/07 | Tue 2/20/07 | |
| 26 |
Define requirements, general problems and issues
|
0% | 1 wk | Mon 1/22/07 | Fri 1/26/07 | |
| 27 |
Add data pool mechanism at BNL, transfer with any method
|
0% | 1 wk | Mon 1/29/07 | Fri 2/2/07 | |
| 28 |
Establish security schem, HPSS auto-synching
|
0% | 1 wk | Mon 2/5/07 | Fri 2/9/07 | |
| 29 |
Test on one or more sites (non-PDSF)
|
0% | 1 wk | Mon 2/12/07 | Fri 2/16/07 | |
| 30 |
Integrate to all participating sites
|
0% | 1 wk | Mon 2/12/07 | Fri 2/16/07 | |
| 31 |
Document data transfer schemeand procedure
|
0% | 2 days | Mon 2/19/07 | Tue 2/20/07 | |
| 32 | ||||||
| 33 |
CVS check-in and cleanup
|
4% | 17 days? | Mon 1/22/07 | Tue 2/13/07 | |
| 34 |
Initial setup, existing framework
|
0% | 17 days | Mon 1/22/07 | Tue 2/13/07 | |
| 35 |
Define proper CVS location for perl, libs, macros
|
0% | 1 day | Mon 1/22/07 | Mon 1/22/07 | Jerome Lauret[10%],Andrew Rose[10%],Lee Barnby[10%] |
| 36 |
Add existing QA macros to CVS
|
0% | 1 day | Tue 1/23/07 | Tue 1/23/07 | Andrew Rose[20%] |
| 37 |
Checkout and test on +1 site (non-PDSF), adjust as necessary
|
0% | 1 wk | Wed 1/24/07 | Tue 1/30/07 | Lee Barnby[10%] |
| 38 |
Bootstrap on +1 site / remove ALL site specifics references
|
0% | 1 wk | Wed 1/31/07 | Tue 2/6/07 | Cristina Suarez[10%] |
| 39 |
Commit to CVS, verify new scripts on all sites, final adjustments
|
0% | 1 wk | Wed 2/7/07 | Tue 2/13/07 | Cristina Suarez[10%],Andrew Rose[10%],Lee Barnby[10%] |
| 40 |
QA and nightly tests
|
17% | 7 days? | Mon 1/22/07 | Tue 1/30/07 | |
| 41 |
Establish a QA area in CVS
|
100% | 1 day? | Mon 1/22/07 | Mon 1/22/07 | |
| 42 |
Check existing QA suite
|
0% | 1 wk | Wed 1/24/07 | Tue 1/30/07 | |
| 43 | ||||||
| 44 |
Development
|
0% | 62 days | Mon 1/22/07 | Tue 4/17/07 | |
| 45 |
General QA consolidation
|
0% | 10 days | Wed 1/31/07 | Tue 2/13/07 | |
| 46 |
Gather feedback from PWG, add QA tests relevant to Physics topics
|
0% | 2 wks | Wed 1/31/07 | Tue 2/13/07 | |
| 47 |
Establish nightly test framework at BNL for embedding
|
0% | 1 wk | Wed 1/31/07 | Tue 2/6/07 | |
| 48 |
General improvements
|
0% | 35 days | Mon 1/22/07 | Fri 3/9/07 | |
| 49 |
Requirements study for an embedding request interface
|
0% | 2 wks | Mon 1/29/07 | Fri 2/9/07 | Andrew Rose[10%],Jerome Lauret[10%] |
| 50 |
Develop new embedding request form compatible with Drupal module
|
0% | 4 wks | Mon 2/12/07 | Fri 3/9/07 | Andrew Rose[10%] |
| 51 |
Test new interface, import old tasks (historical purposes)
|
0% | 5 days | Mon 1/22/07 | Fri 1/26/07 | Andrew Rose[10%],Cristina Suarez[10%] |
| 52 |
Distributed Computing
|
0% | 20 days | Wed 2/14/07 | Tue 3/13/07 | |
| 53 |
Use SUMS framework to submit embedding, establish first XML
|
0% | 1 wk | Wed 2/14/07 | Tue 2/20/07 | Lee Barnby[10%] |
| 54 |
Test on one site
|
0% | 1 wk | Wed 2/21/07 | Tue 2/27/07 | Lee Barnby[10%] |
| 55 |
Test on all sites, adjust as necessary
|
0% | 2 wks | Wed 2/28/07 | Tue 3/13/07 | Cristina Suarez[10%],Andrew Rose[10%],Lee Barnby[10%] |
| 56 |
Gridfication
|
0% | 25 days | Wed 3/14/07 | Tue 4/17/07 | |
| 57 |
Test XML using Grid policy (one site)
|
0% | 1 wk | Wed 3/14/07 | Tue 3/20/07 | |
| 58 |
Establish test of data transfer method, GSI enabled HPSS access possible
|
0% | 1 wk | Wed 3/21/07 | Tue 3/27/07 | |
| 59 |
Regression and stress test on one site
|
0% | 1 wk | Wed 3/28/07 | Tue 4/3/07 | |
| 60 |
Test on +1 site, infrastructure consolidation
|
0% | 2 wks | Wed 4/4/07 | Tue 4/17/07 | |
| 61 | ||||||
| 62 |
Embedding operation
|
25% | 261 days? | Mon 1/1/07 | Mon 12/31/07 | |
| 63 |
PDSF support
|
50% | 261 days? | Mon 1/1/07 | Mon 12/31/07 | Andrew Rose[10%] |
| 64 |
BHAM support
|
10% | 261 days? | Mon 1/1/07 | Mon 12/31/07 | Lee Barnby[10%] |
| 65 |
UIC Support including QA
|
15% | 261 days? | Mon 1/1/07 | Mon 12/31/07 | Olga Barranikova[5%],Cristina Suarez[10%] |
Chain check, momentum issues
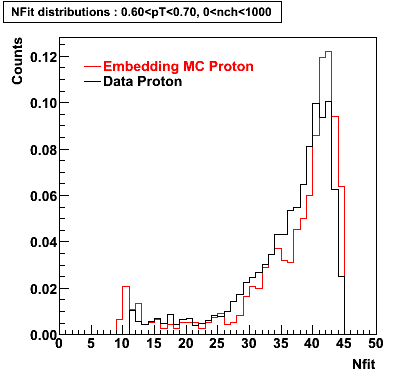
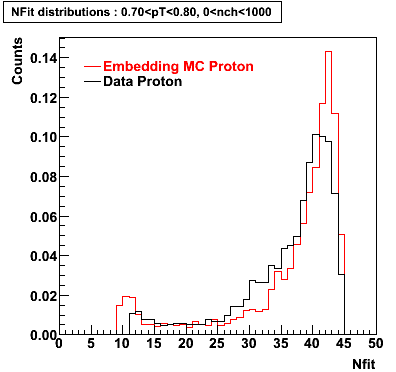
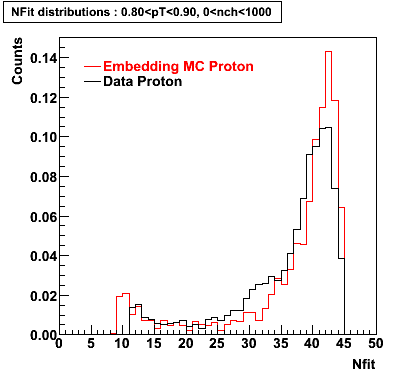
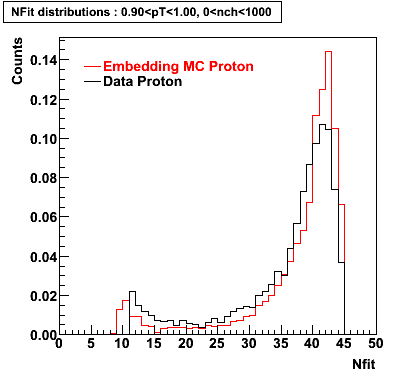
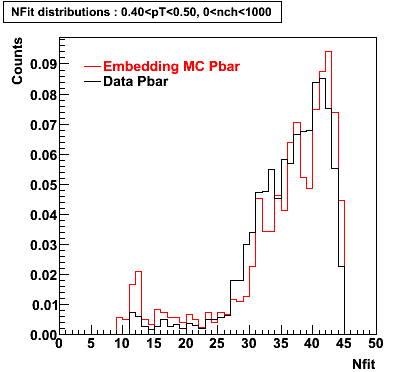
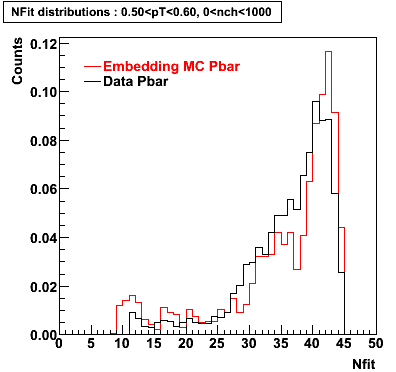
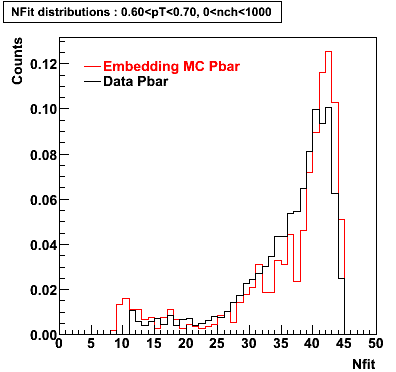
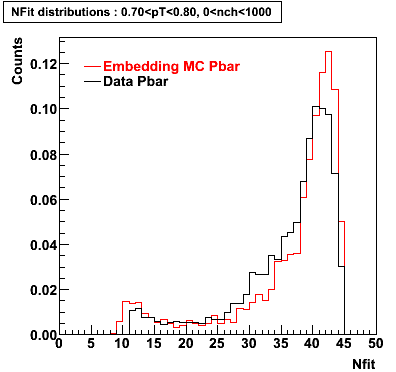
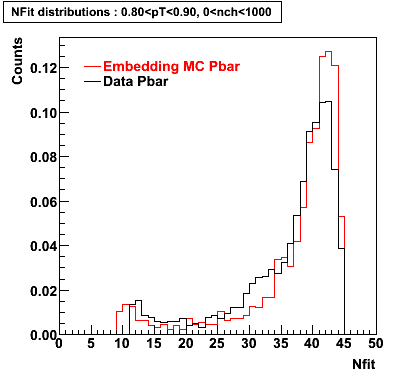
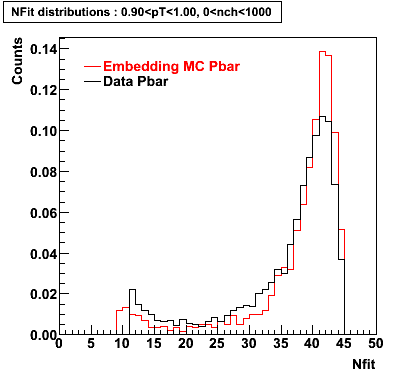
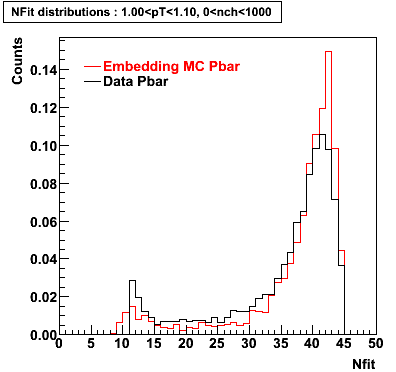
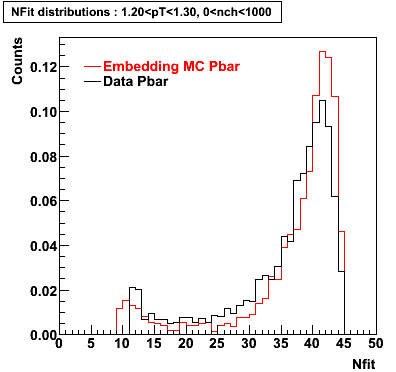
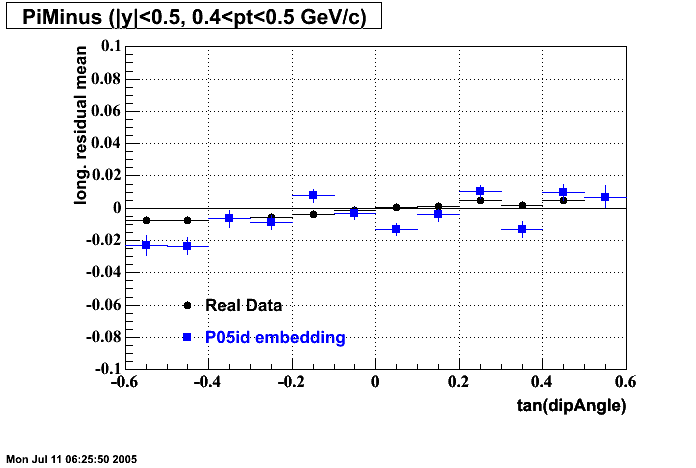
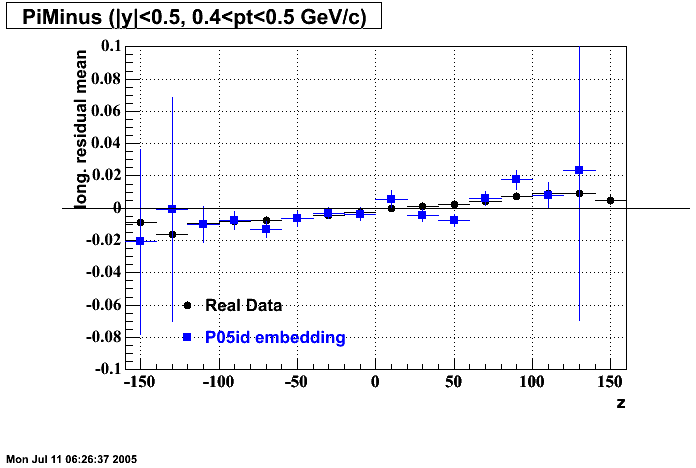
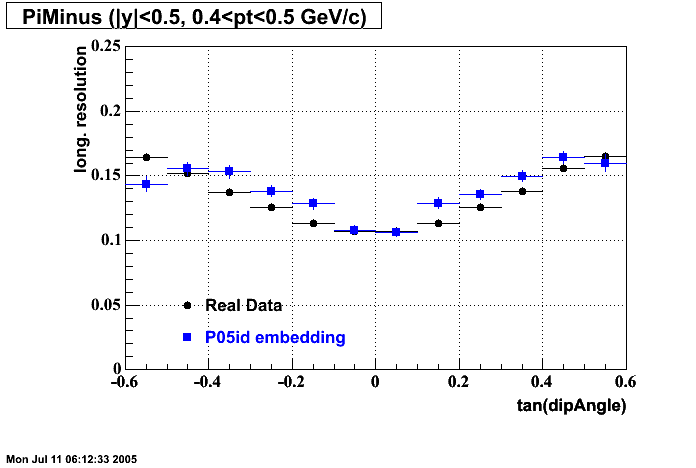
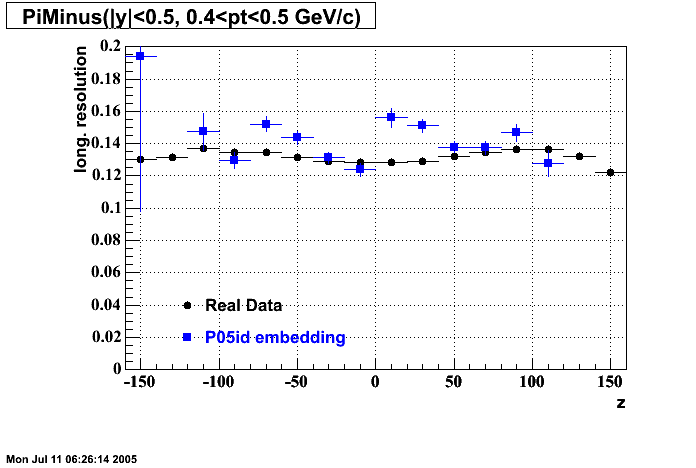
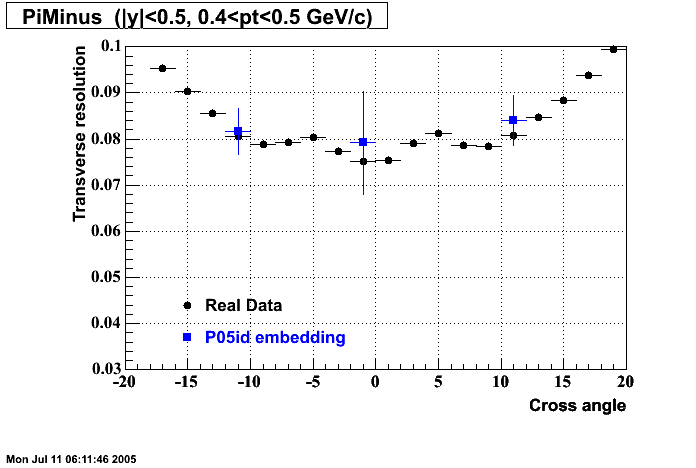
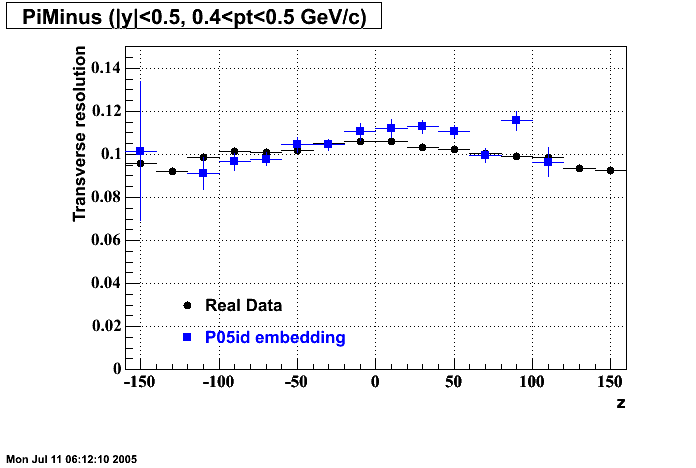
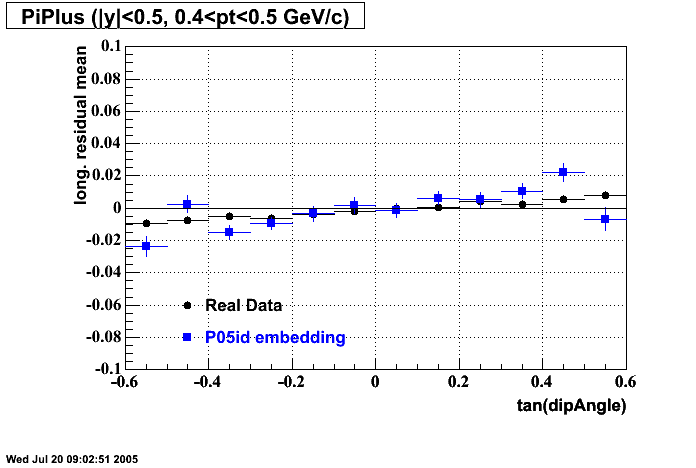
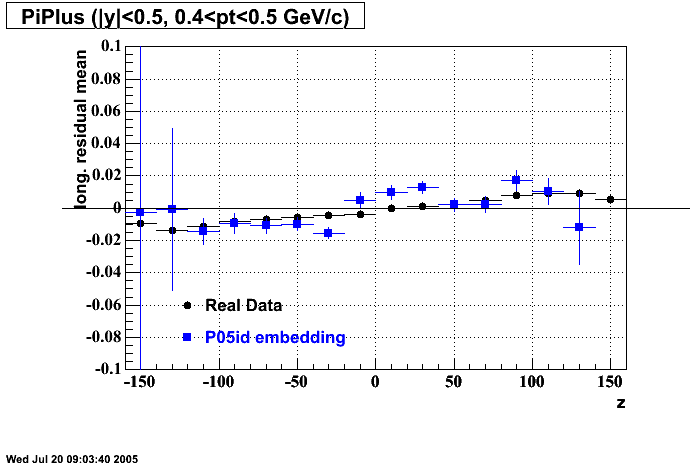
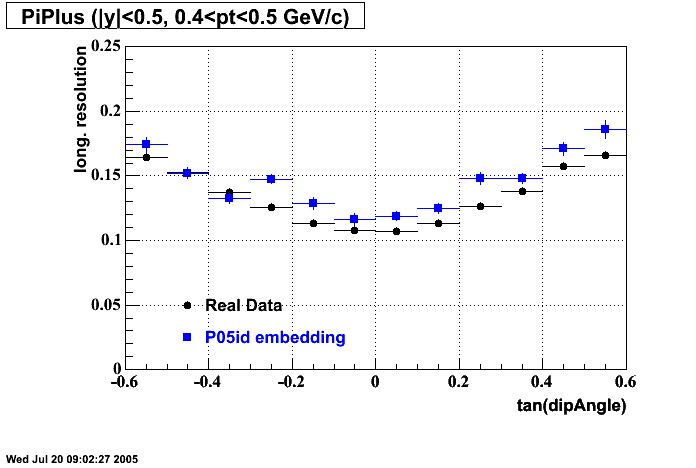
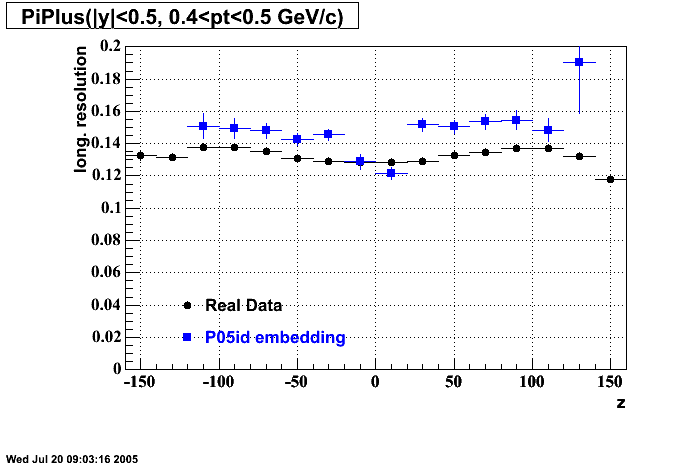
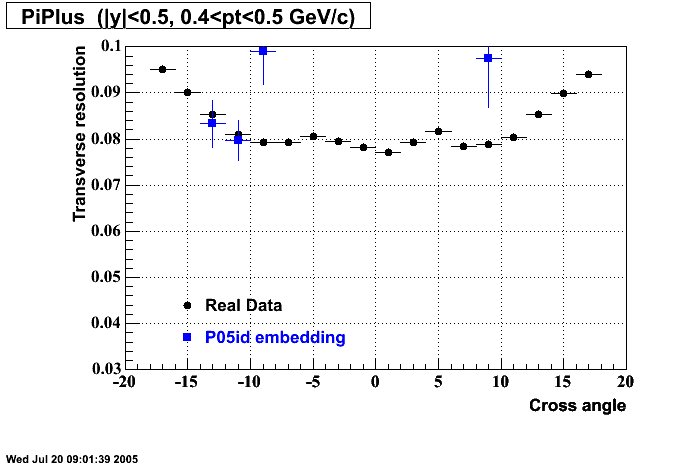
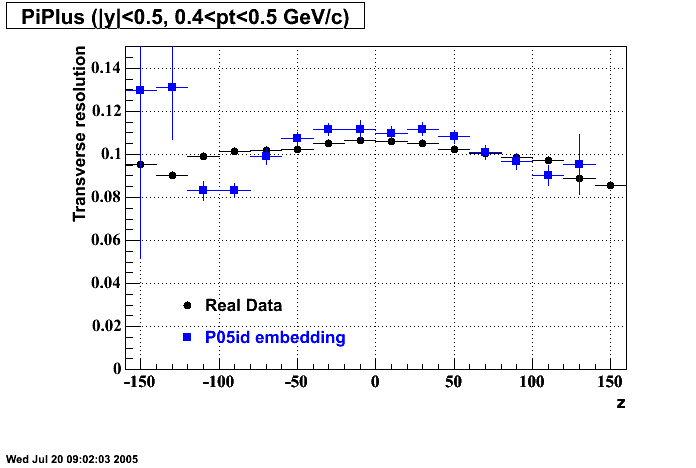
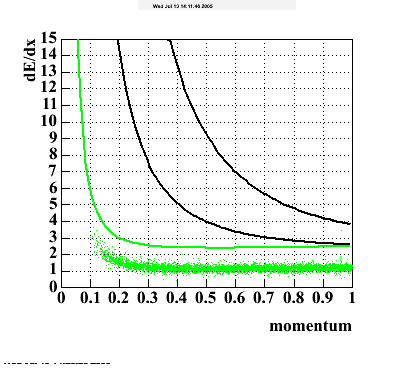
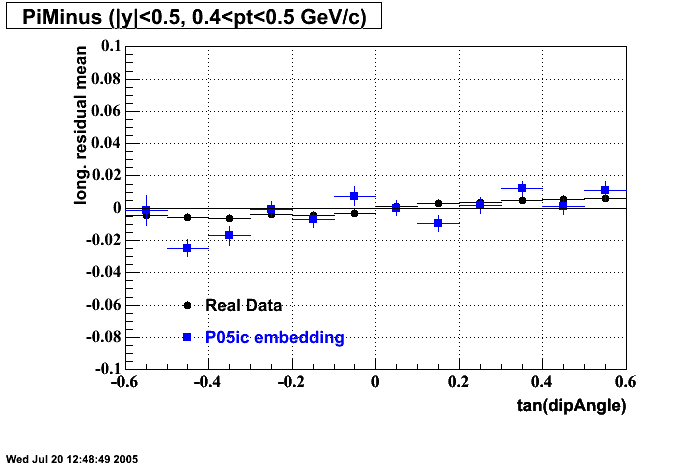
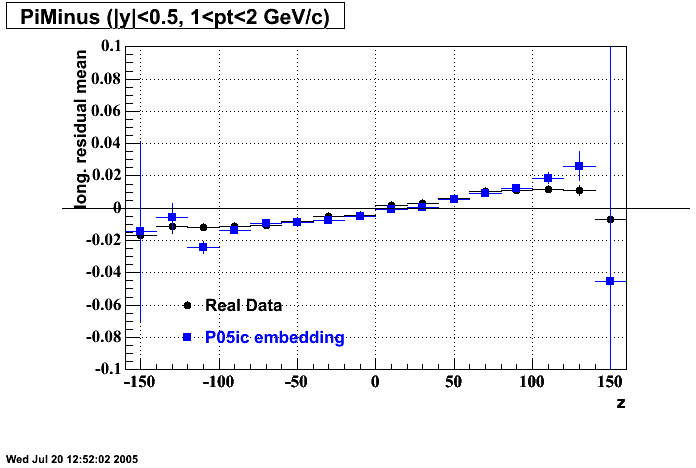
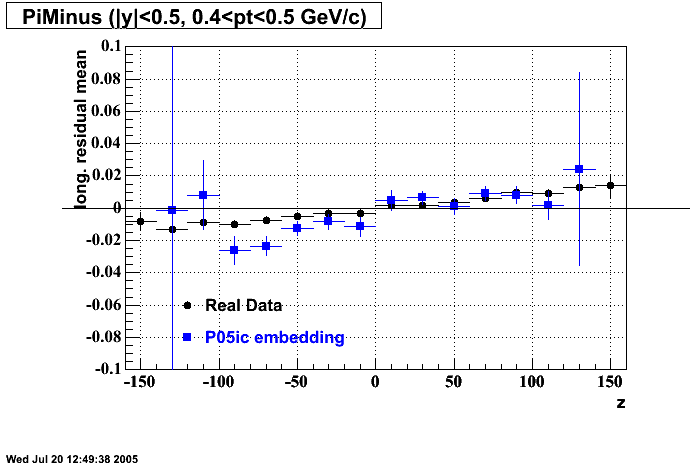
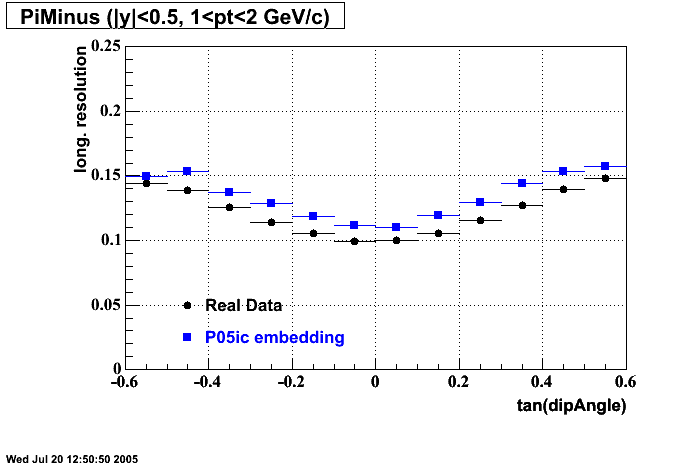
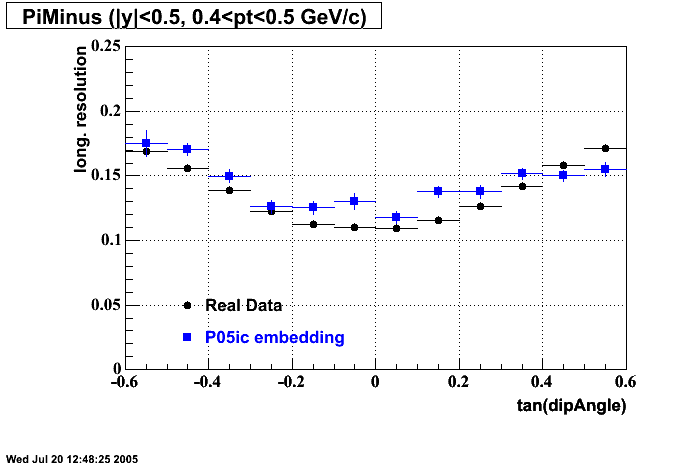
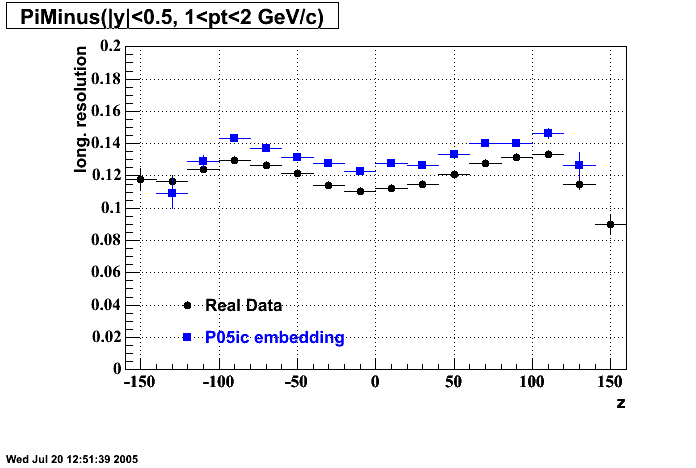
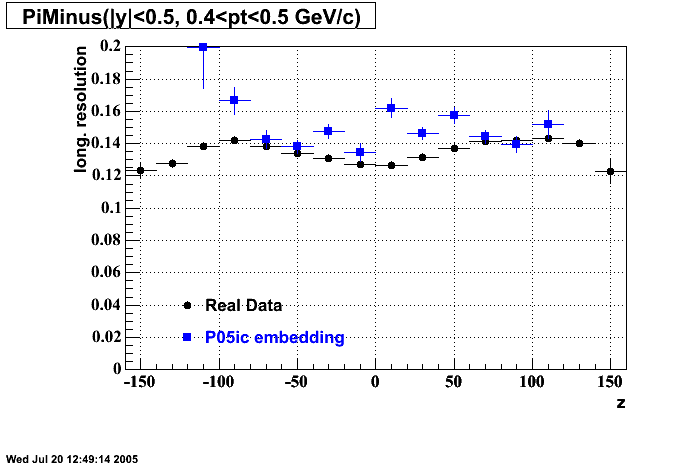
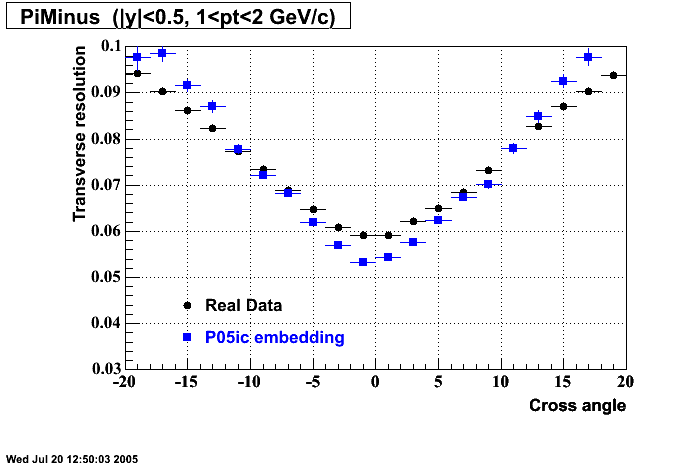
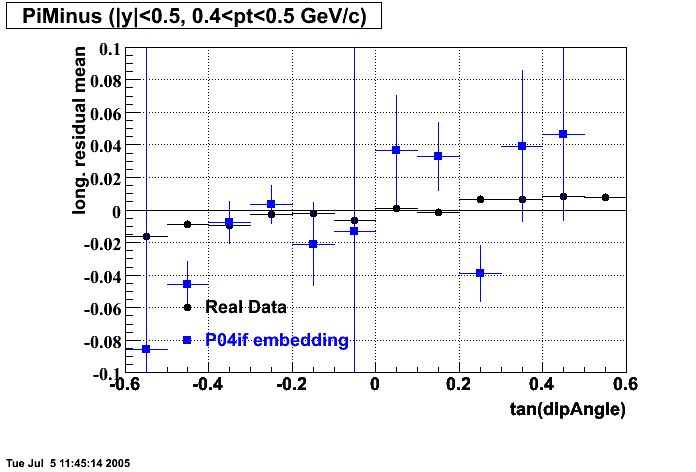
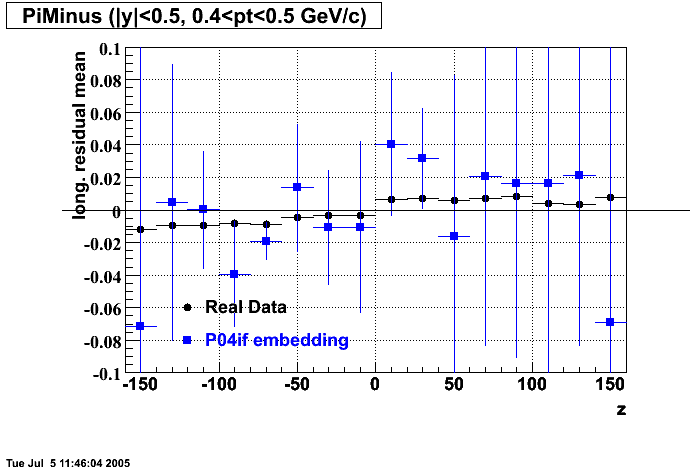
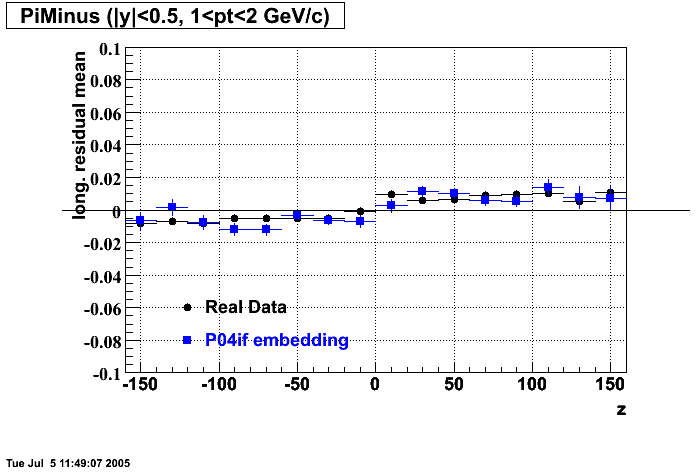
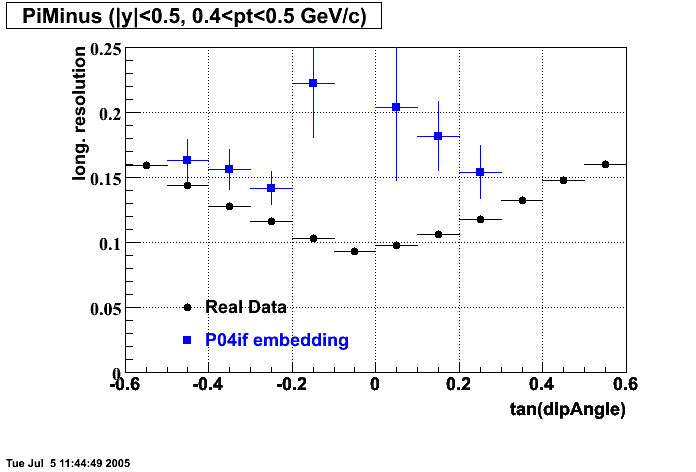
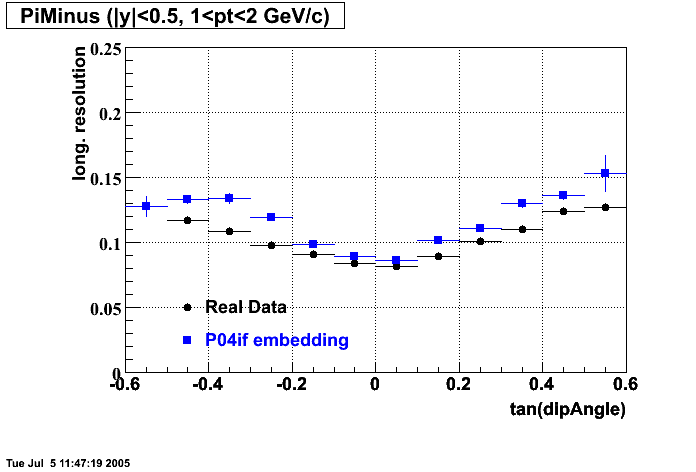
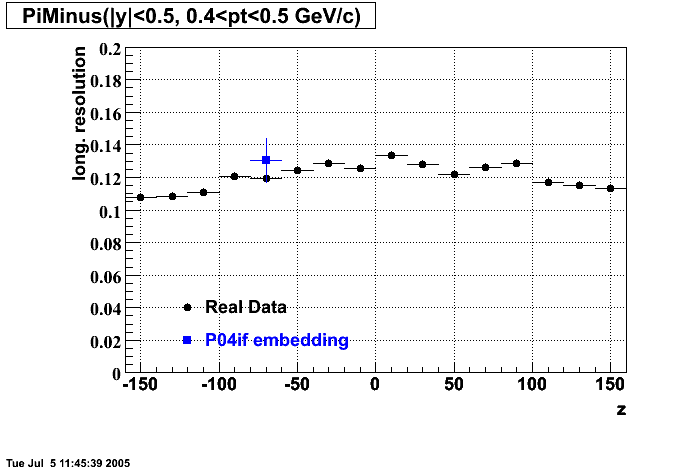
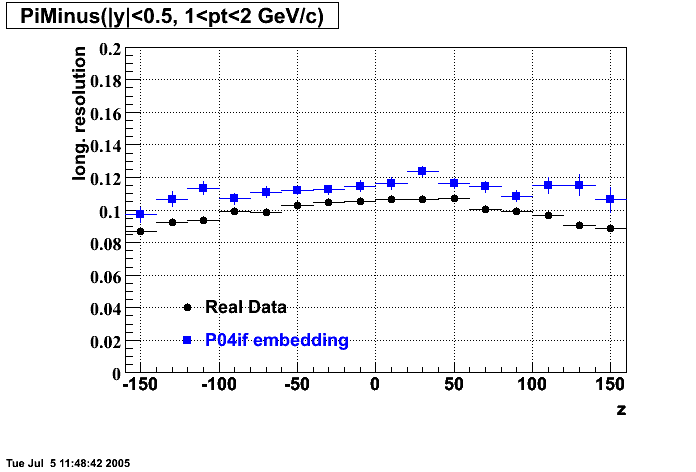
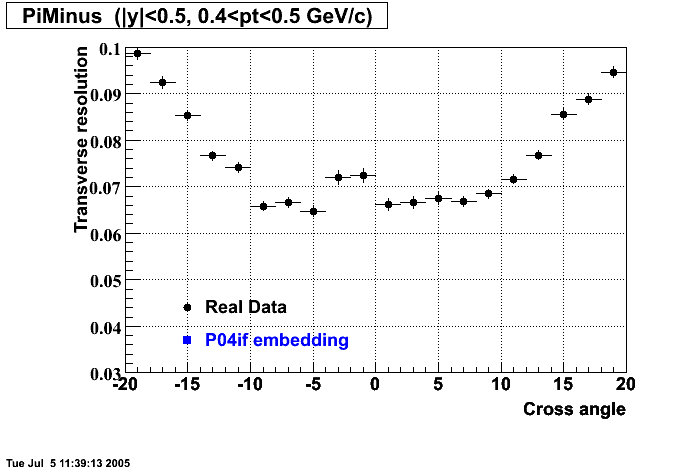
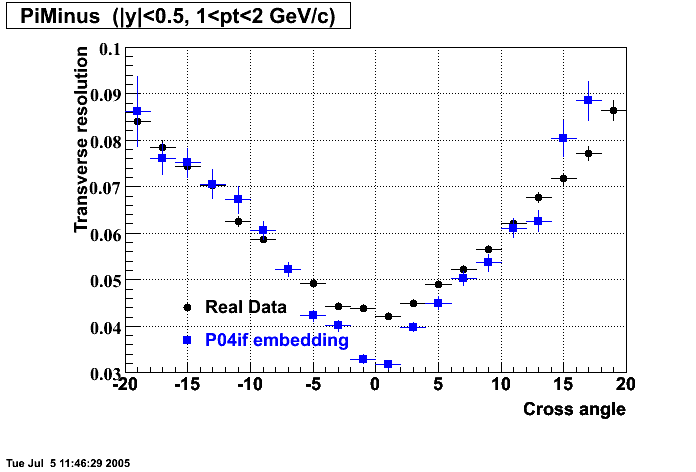
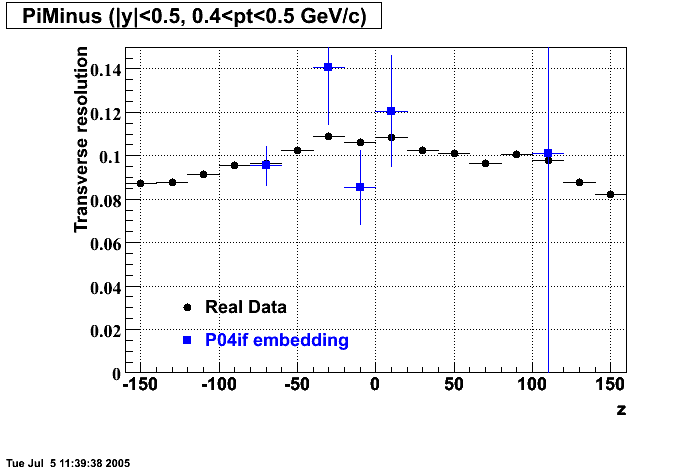
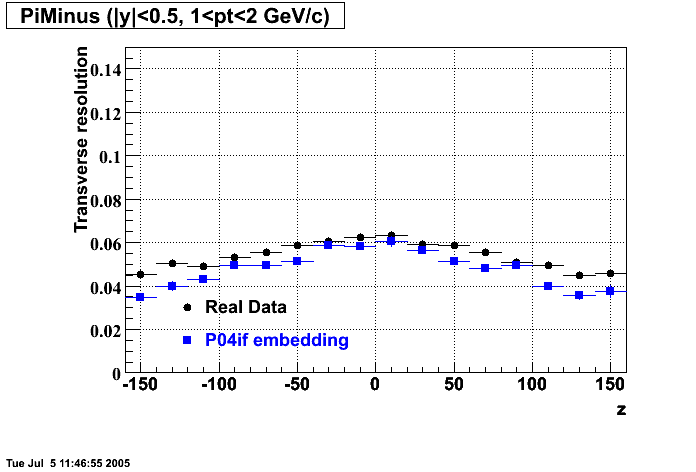
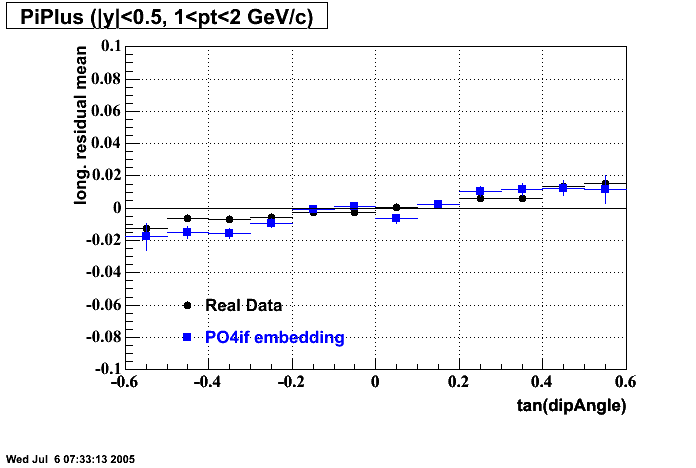
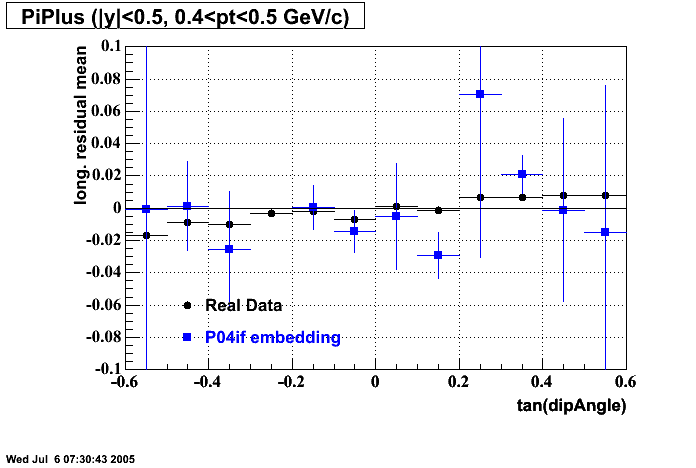
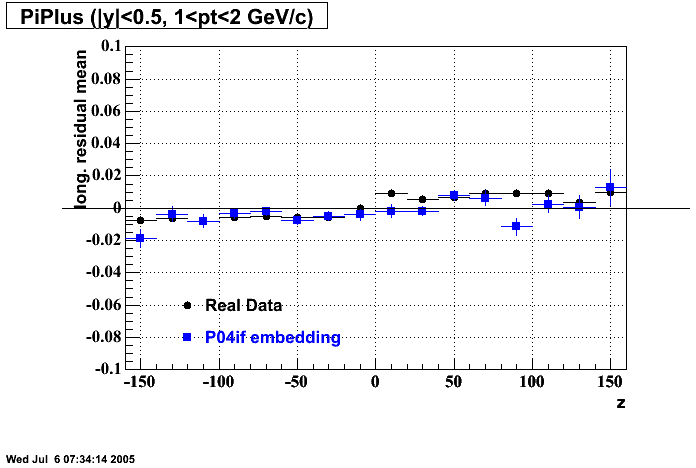
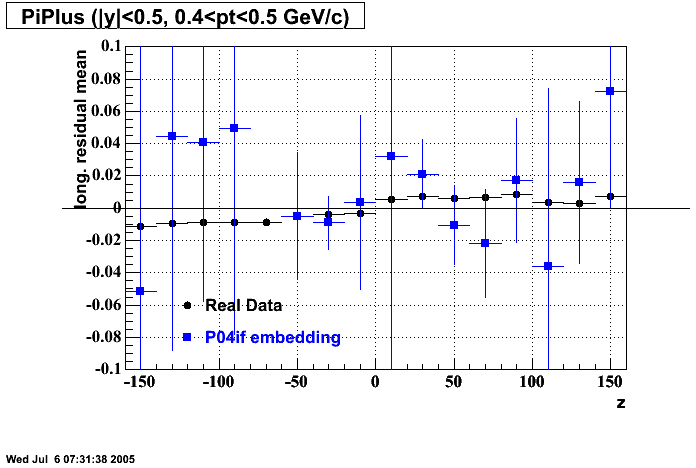
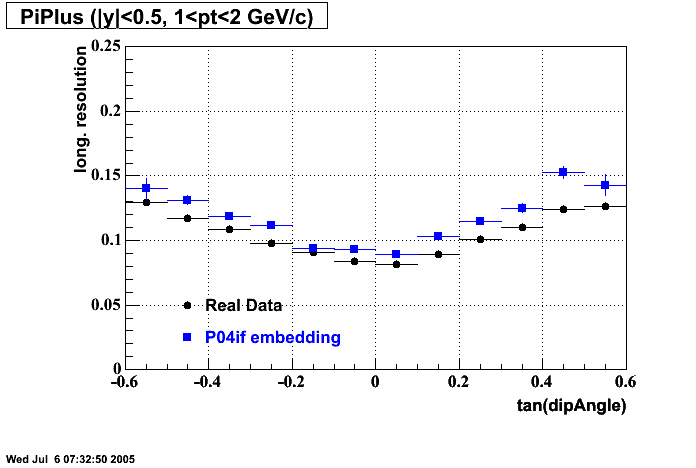
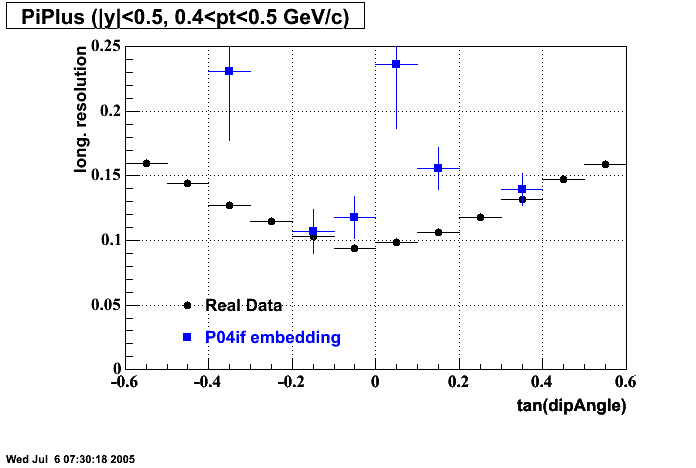
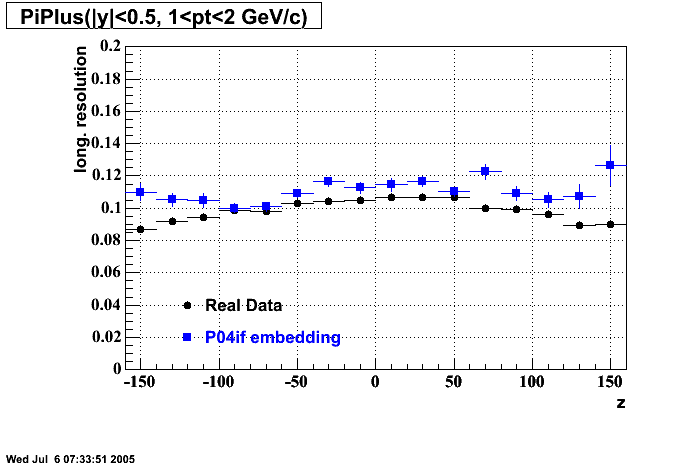
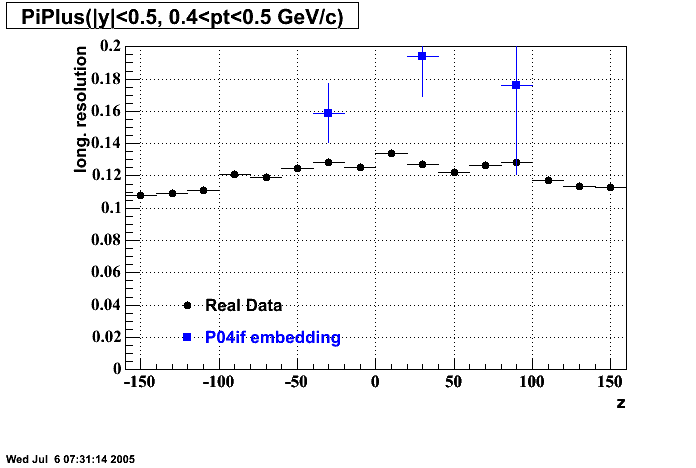
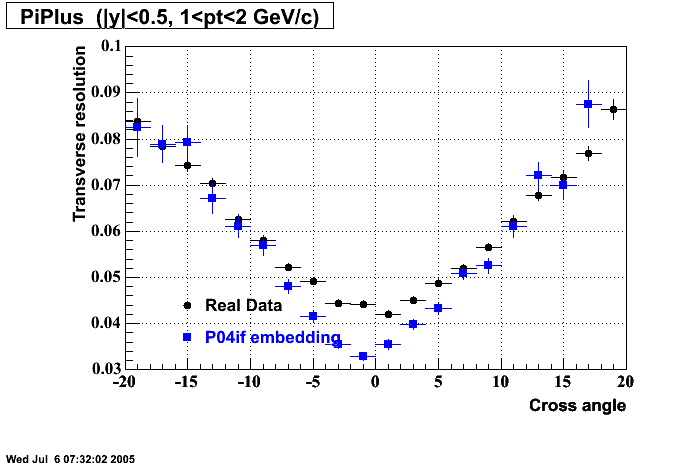
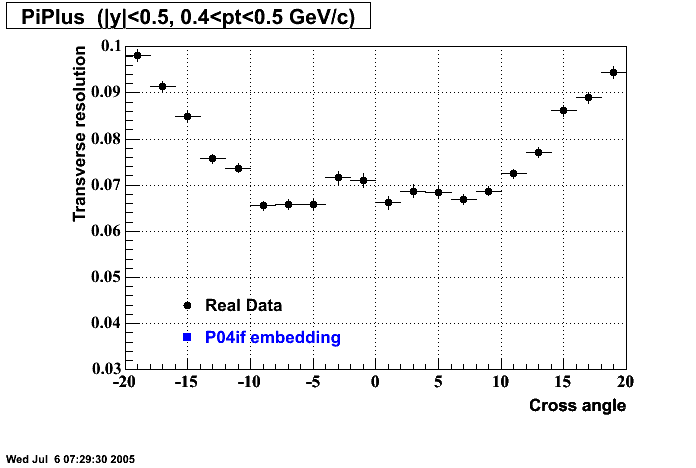
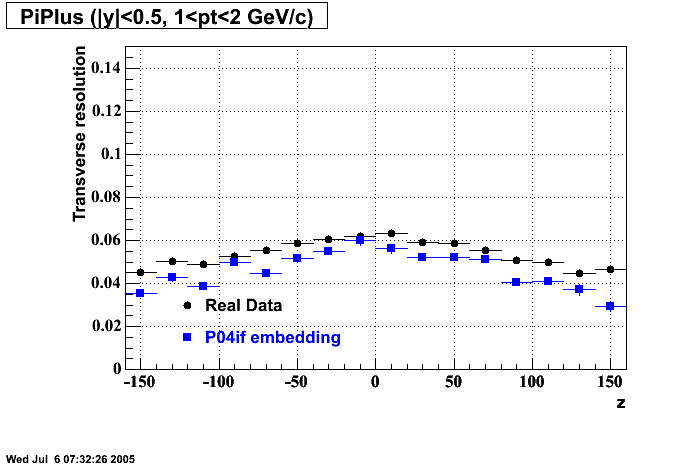
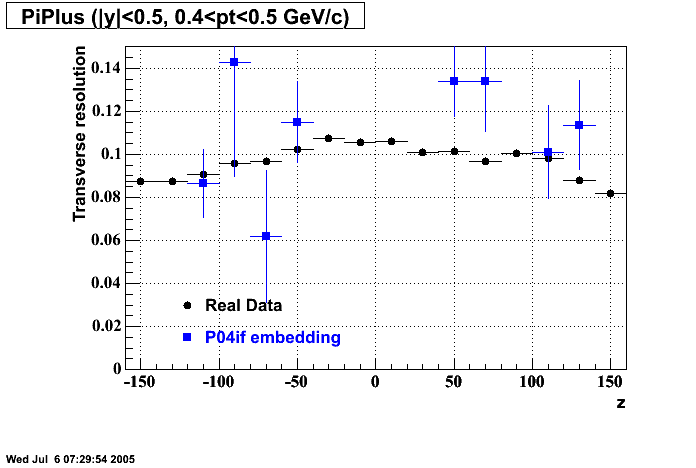
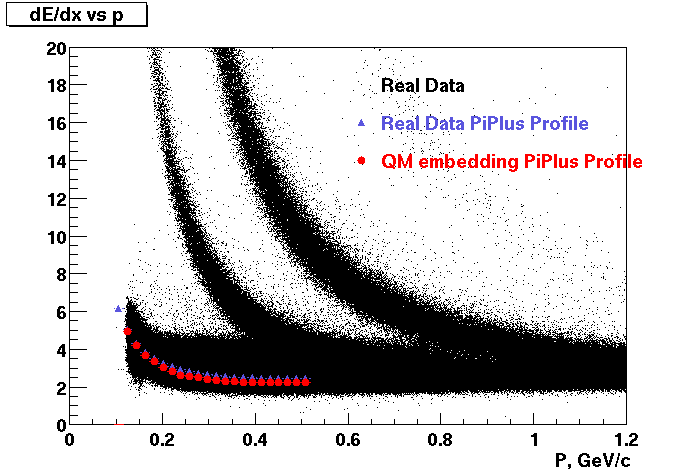
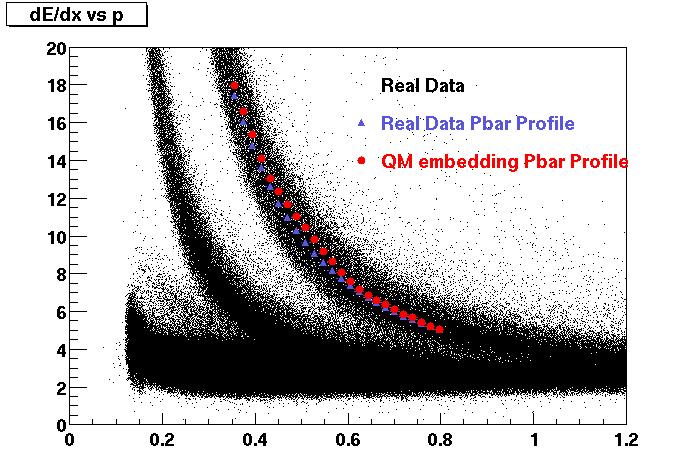
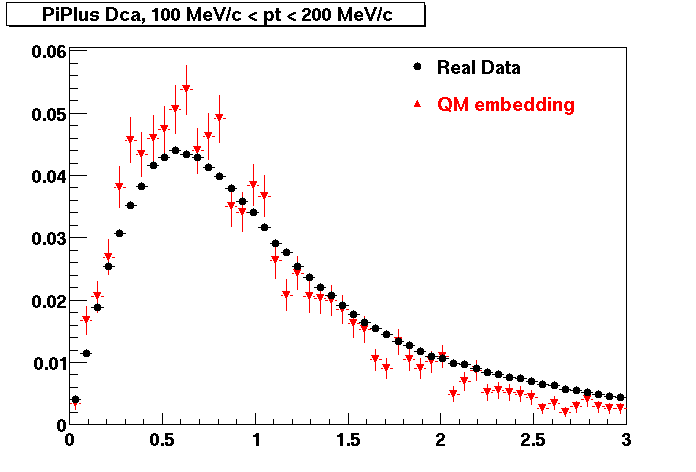
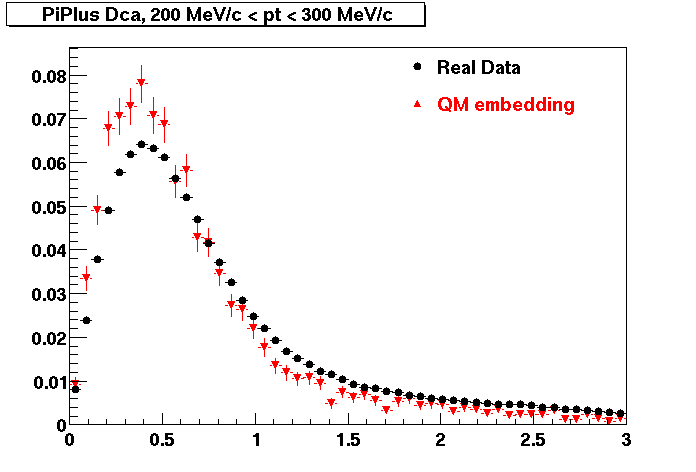
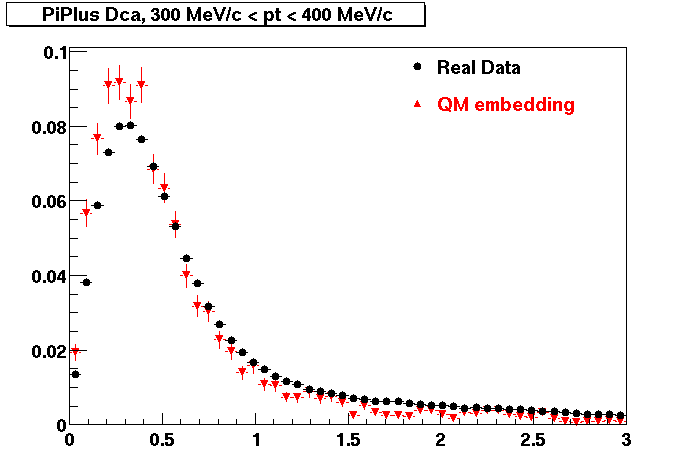
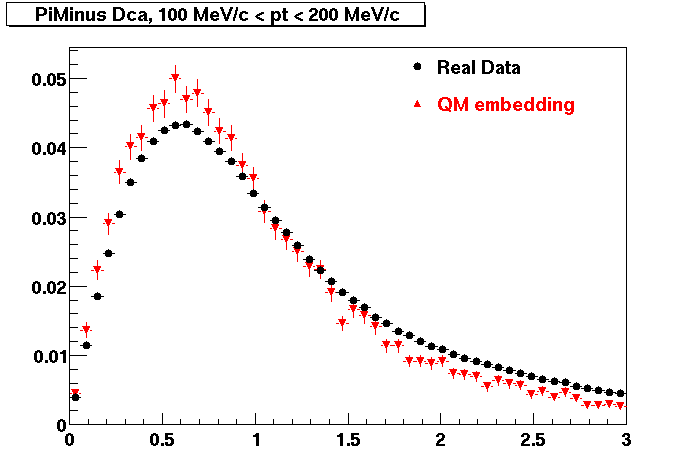
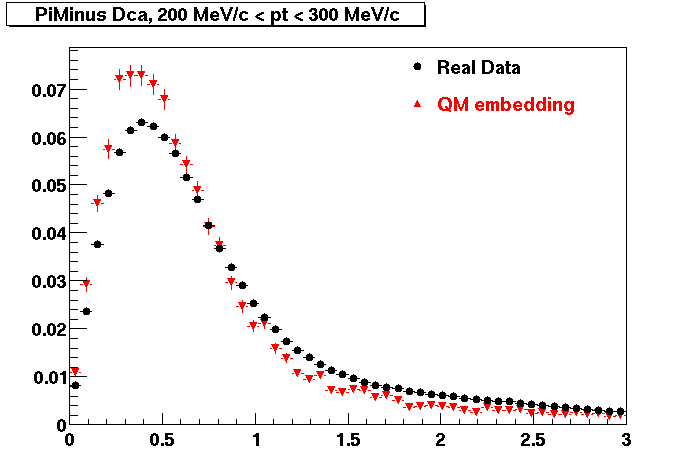
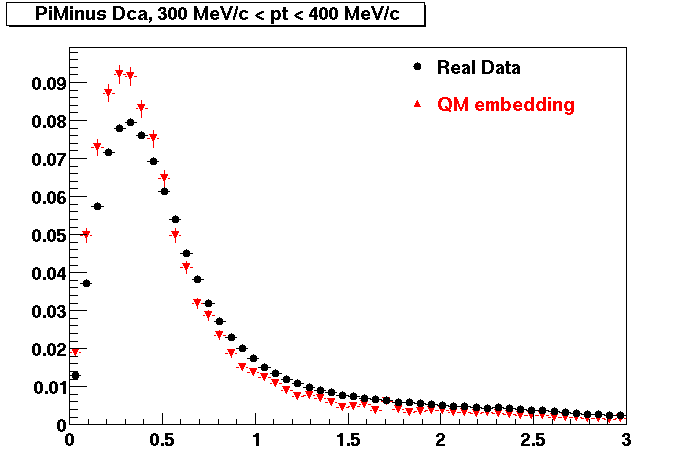
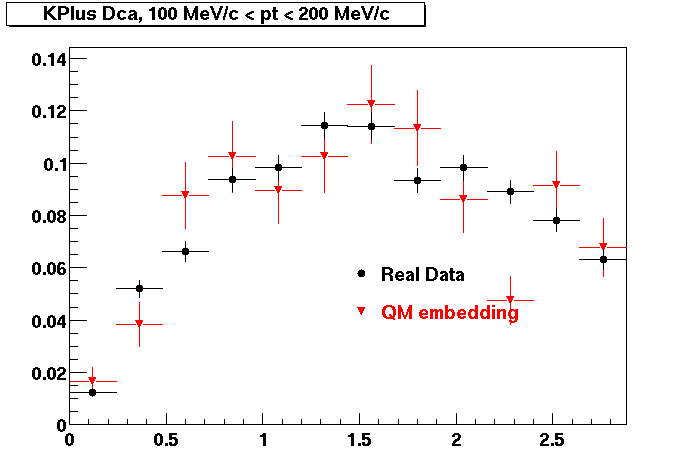
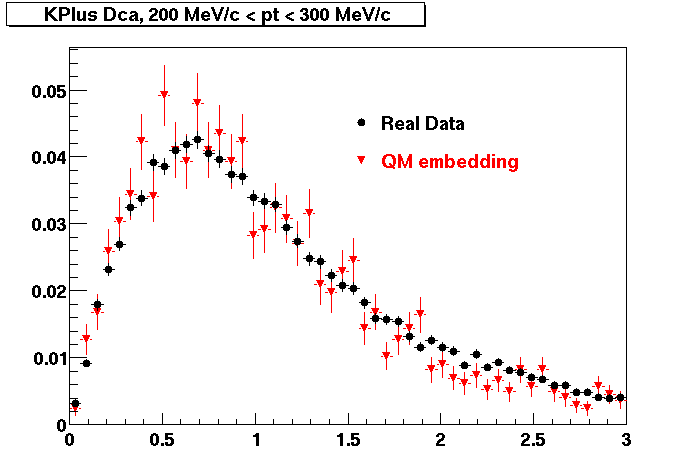
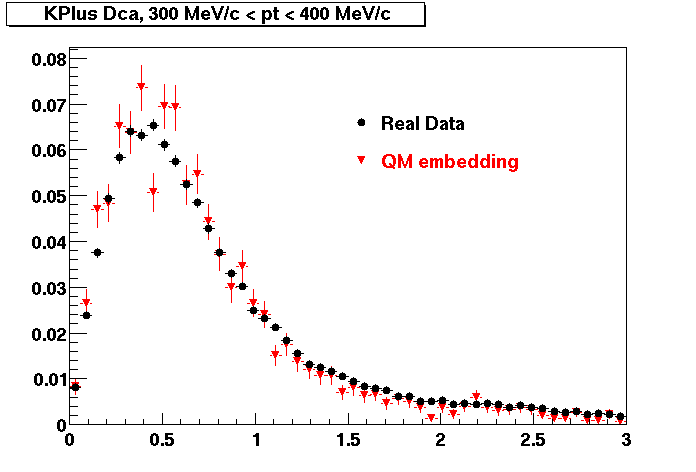
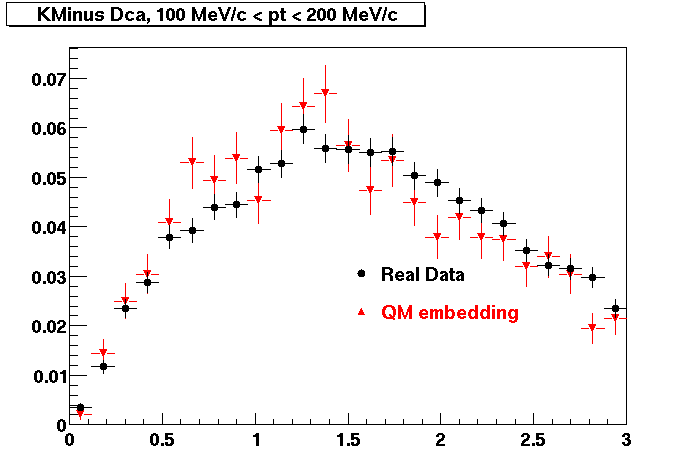
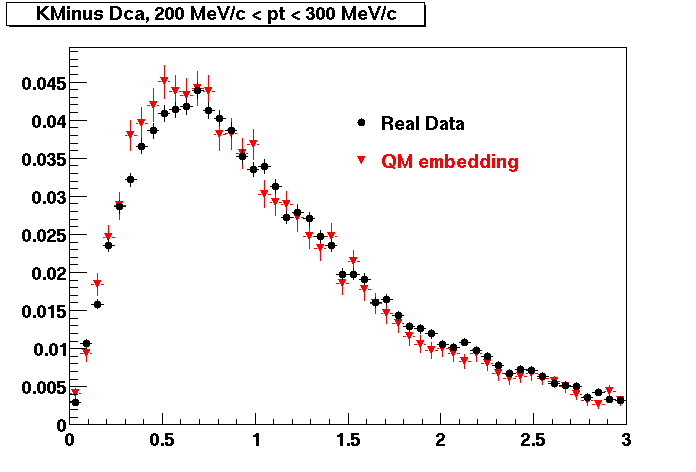
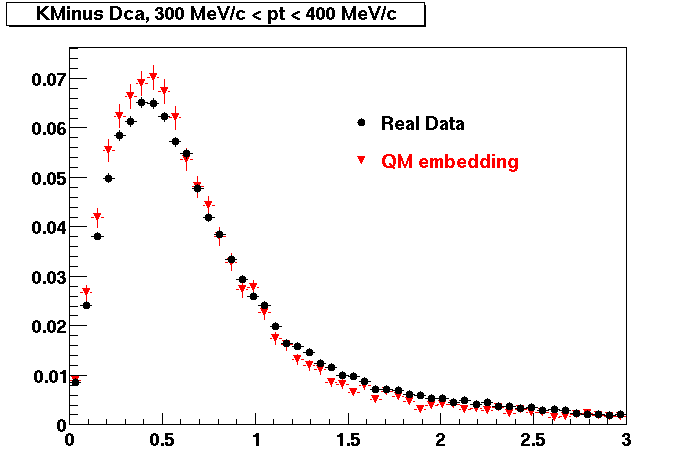
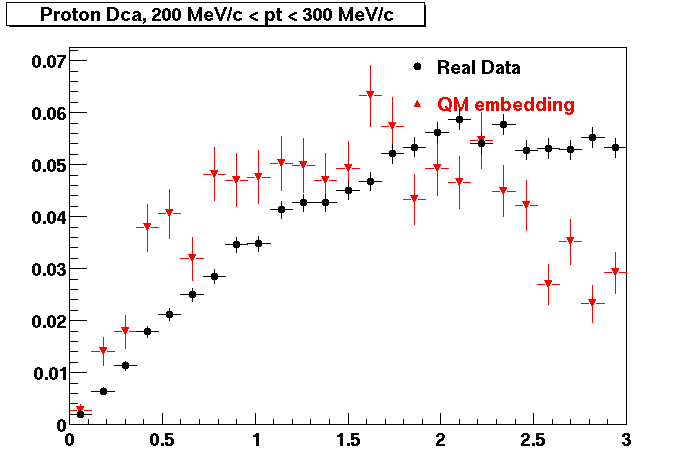
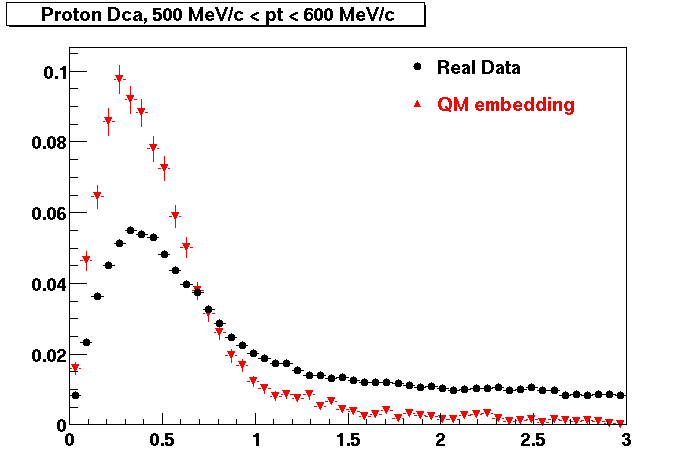
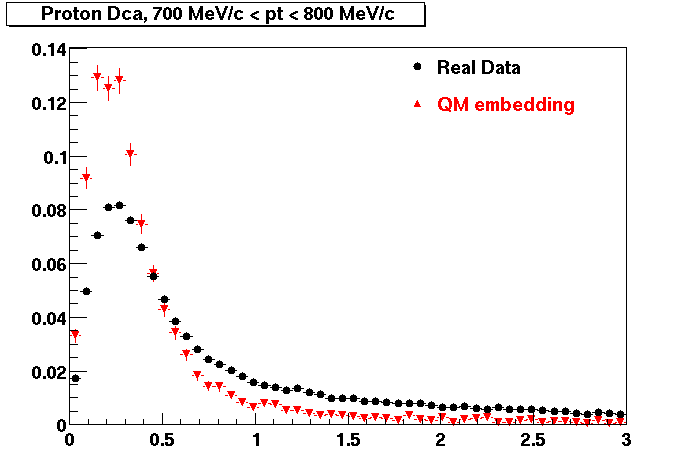
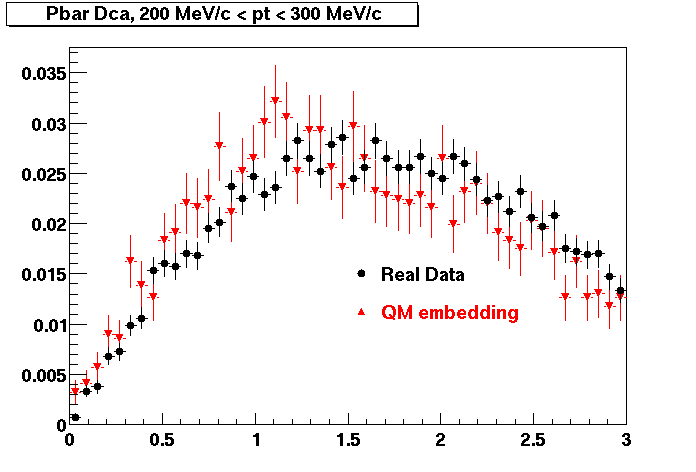
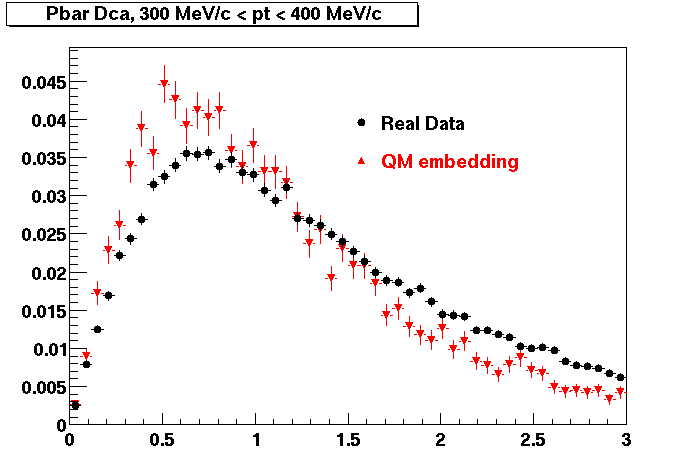
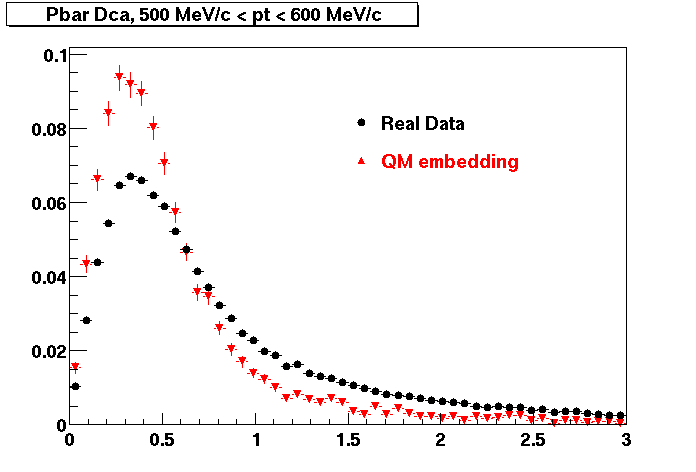
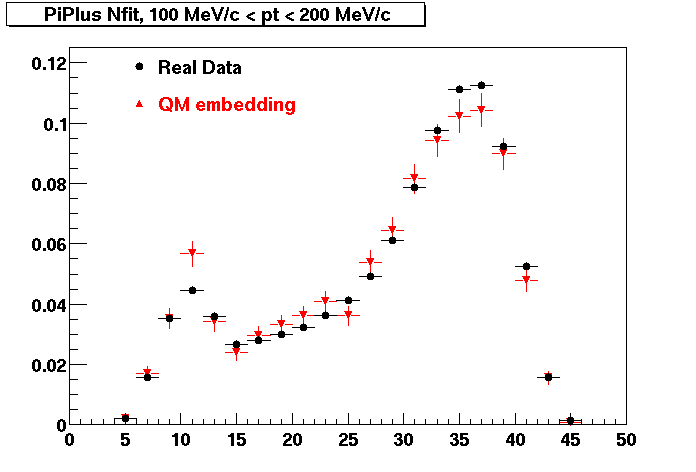
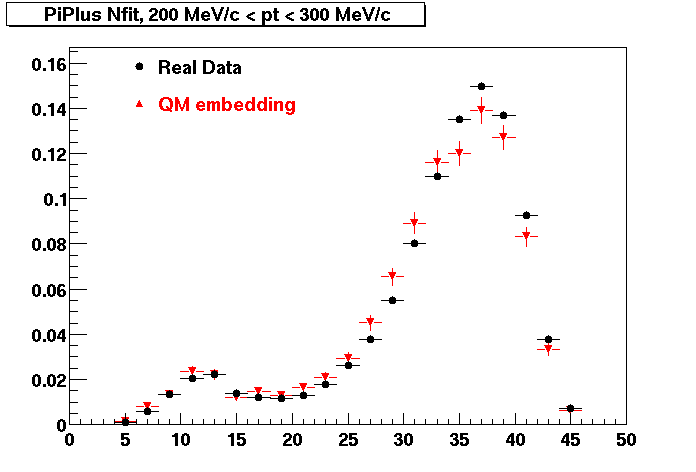
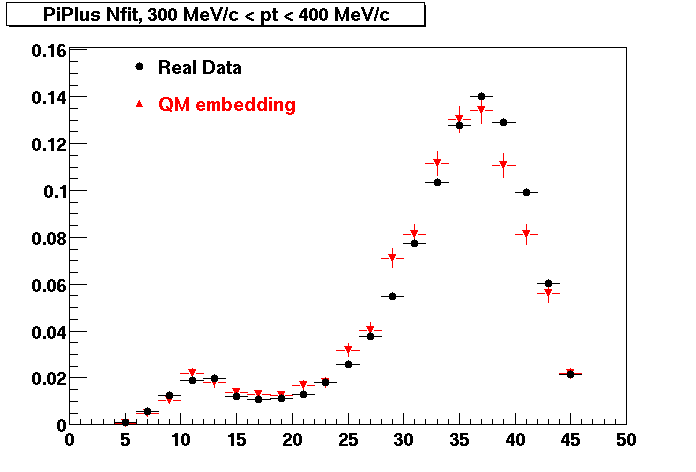
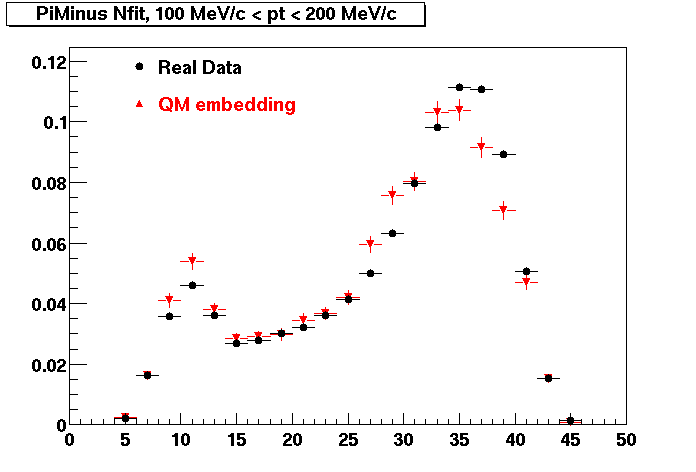
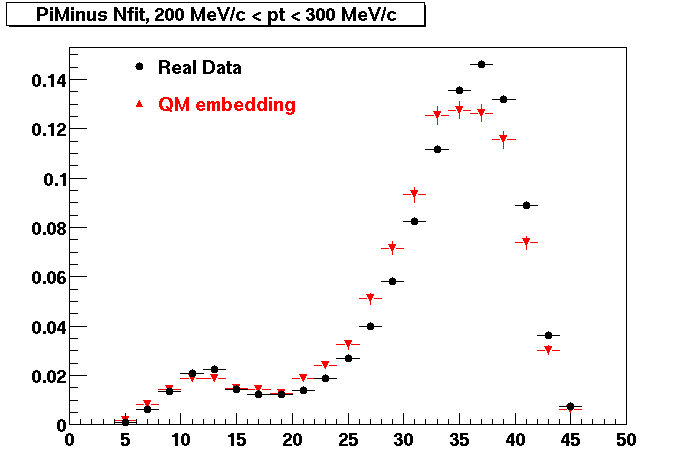
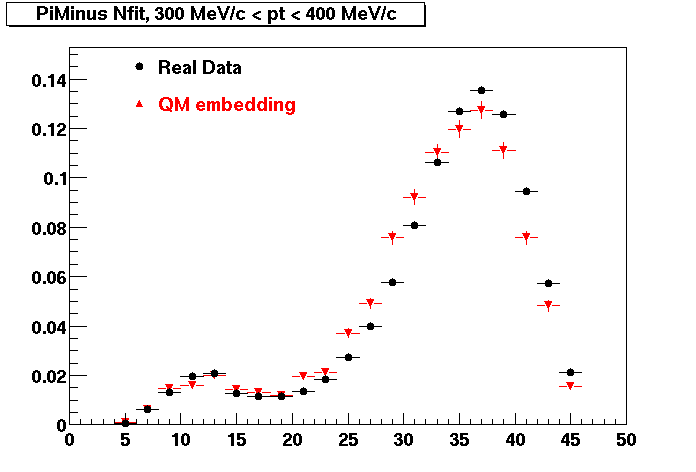
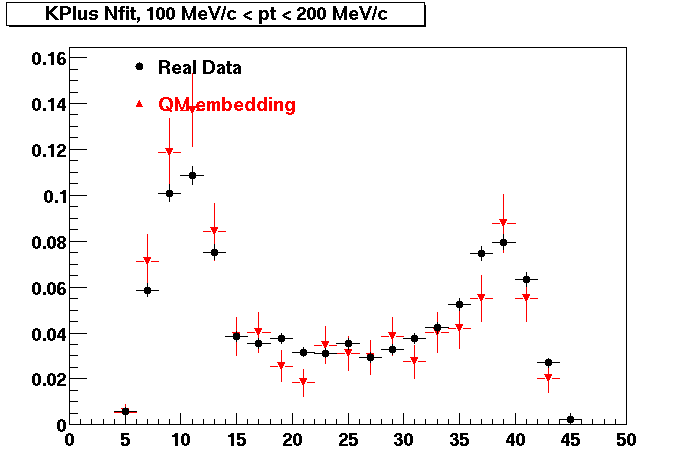
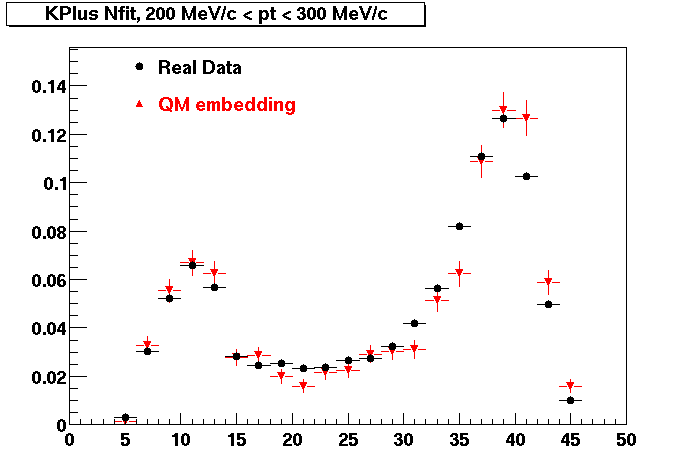
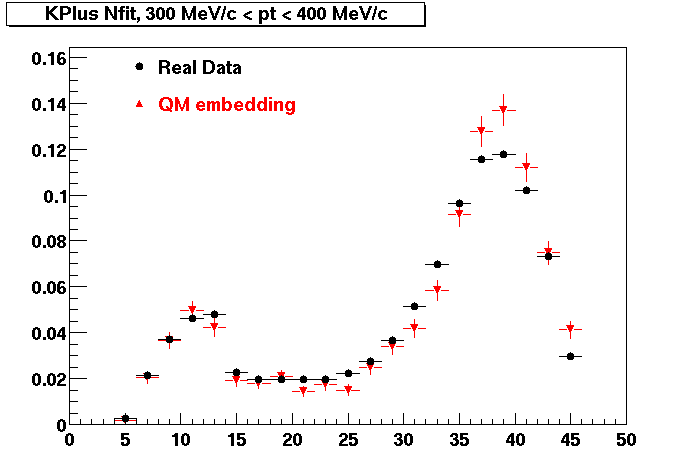
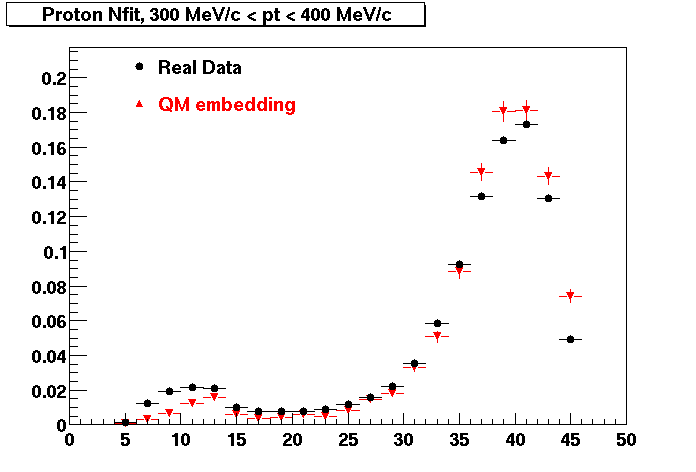
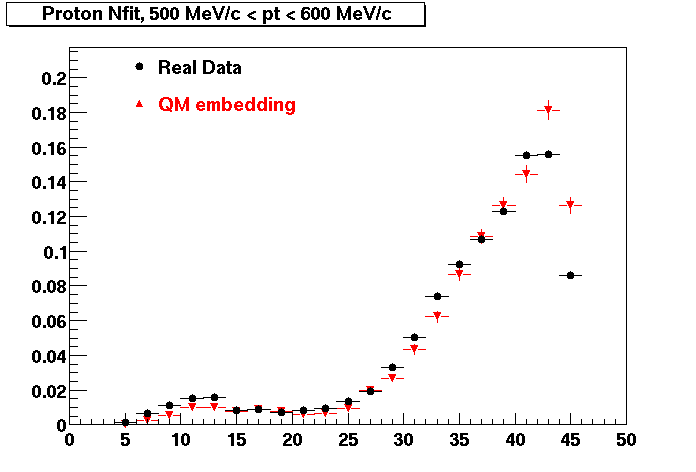
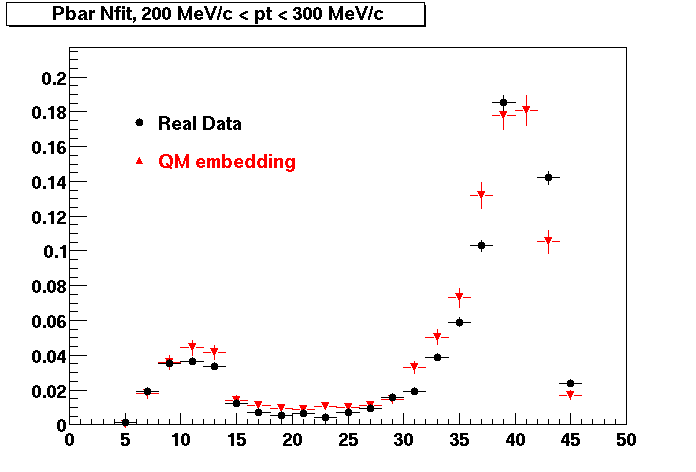
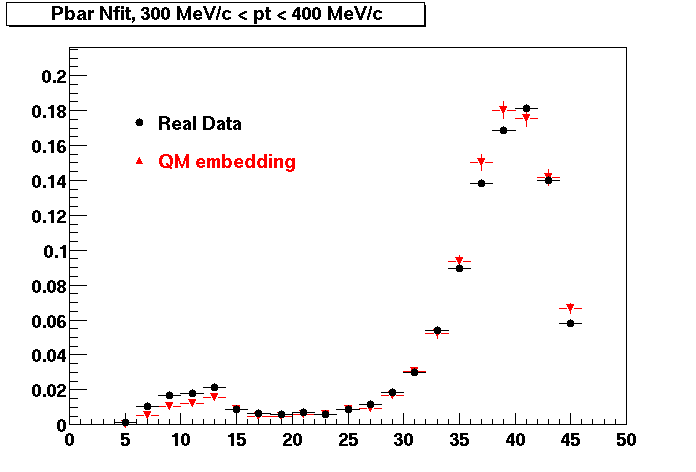
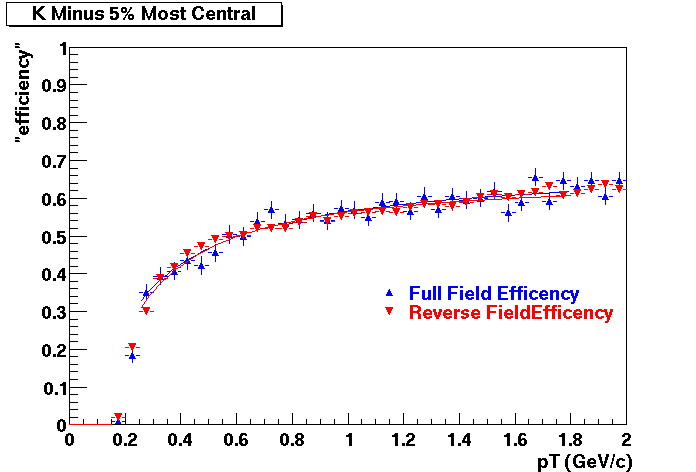
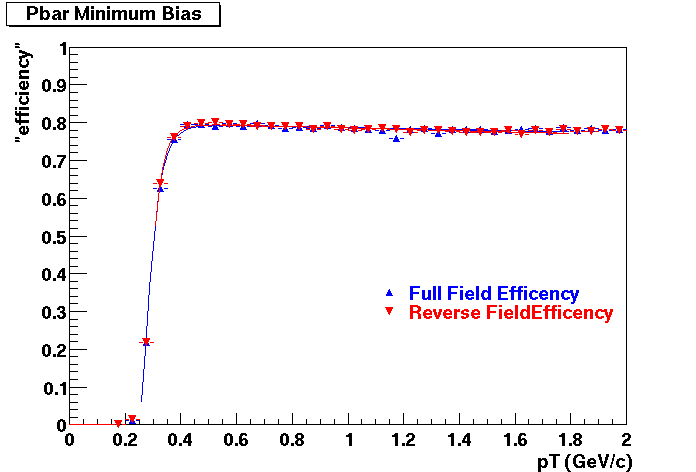
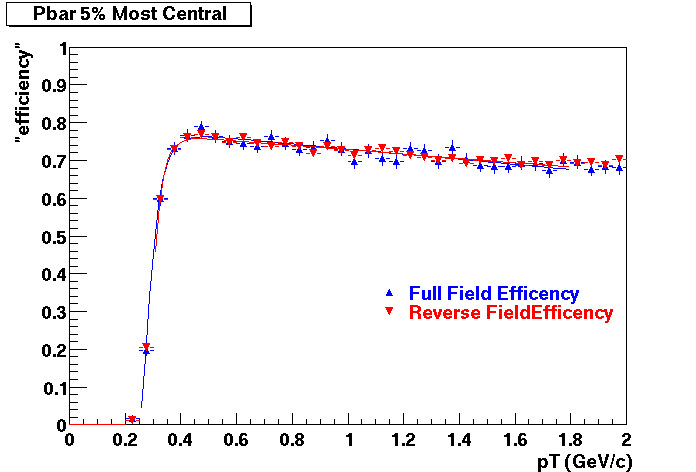
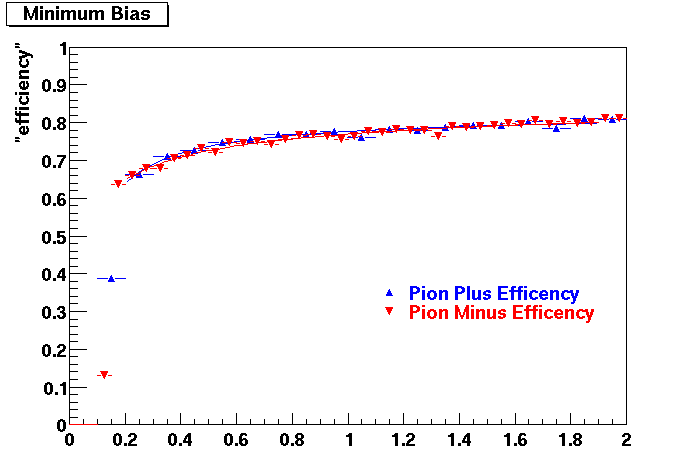
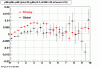
The below plots were the basis for the second You do not have access to view this node as a follow of the You do not have access to view this node.
Olga versus Victor plots - Are they consistent?
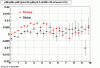
Eloss proximity from Olga
|
Eloss idtruth from Olga
|

Victor's P diff (proximity)
|

Victor's P diff (idtruth)
|
idtruth versus proximity
Victor's proximity plot
|

Victor's idtruth
|
bfcMixer.C Reshape
b>Date: Wednesday, 18 April 2007Time: 12:21:45
Topic: Embedding Reshape
18 April 2007 12:21:56
Talked to Yuri on Monday (16th)
He would like 3 things worked on.
1. Integration of MC generation part into bfcMixer.C
Basically all kumac commands can be done in macro using gstar.
These would become part of "chain one"
Also need to read in a tag or MuDst file to find vertex to use for generating particles.
Can probably see how this works from bfc.C itself as bfc.C(1) creates particles and runs them through reconstruction.
- actually I could not it is inside bfc.C or StBFChain because it is part of St_geant_Maker
2. Change Hit Mover so that it does not move hits derived from MC info (based on ID truth %age)
3. [I forgot what 3 was!]
Rough sketch of chain modifications for #1
Current bfcMixer
(StChain)Chain
(StBFChain)daqChain<--daq file
(StBFChain simChain<--fz file
<---.dat file with vertex positions
MixerMaker
(StBFChain)recoChain
New bfcMixer
(StChain)Chain
(
StBFChain)daqChain<--daq file
(StBFChain)simChain
|
Geant-?-SetGeantMaker<--tags file
MixerMaker
(StBFChain)recoChain
Break down into sub-tasks.
a) Run bfcMixer.C on a daq file with an associated fz and data file (to check that it works!)
b) Ignore fz file and generate MC particle (any!) on the fly
c) reading from tags file generate MC particles at desired vertex & with desired mult.
d) tidy up specify parameter interface (p distn, geant ID etc.)
Embedding Procedures
Overview
This document lists the procedures used for requesting embedding, running embedding jobs, and retrieving data. Please note that some steps require privileged accounts.
Embedding Documentation
Overview
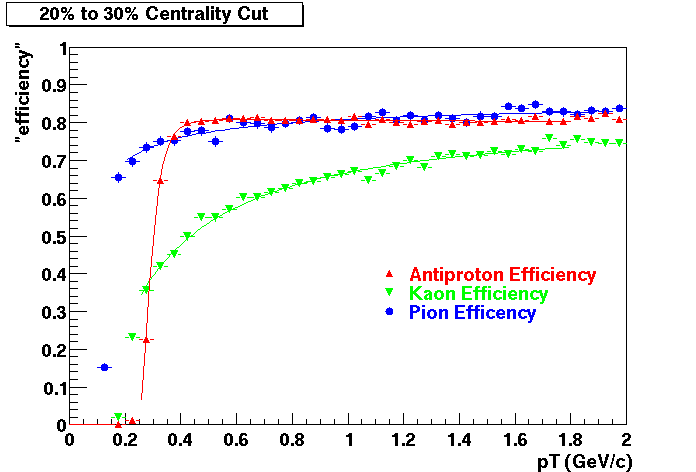
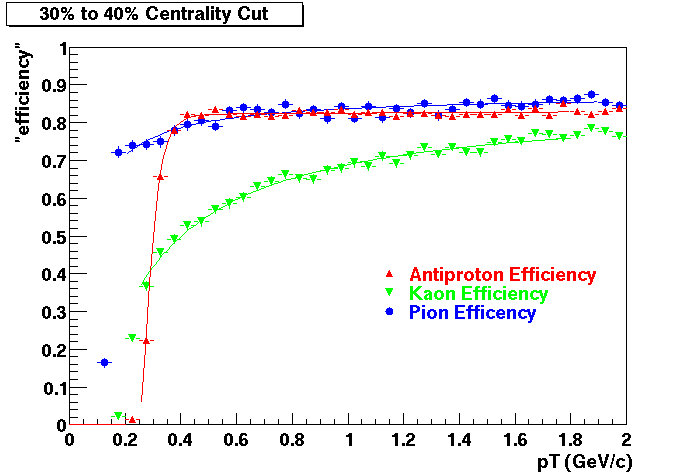
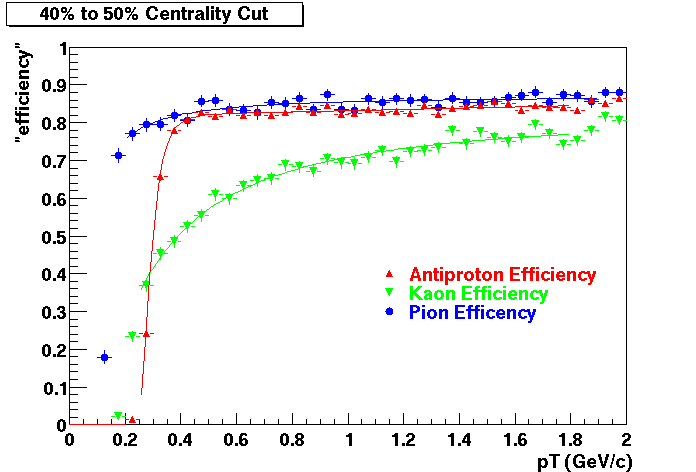
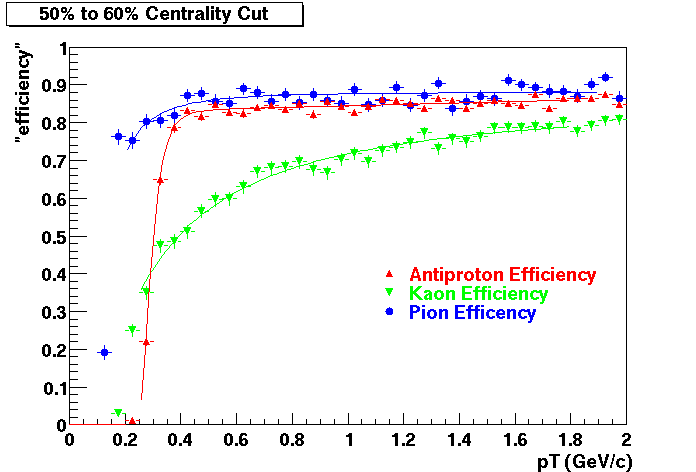
The purpose of embedding is to provide physicists with a known; Monte Carlo tracks, where the kinematics and particle type is known exactly, are introduced into the realistic environment seen in the analysis - a real data event. How reconstruction preforms on these "standard candle" tracks provides a baseline which can be used to correct for acceptance and effciency effects in various analyses.
In STAR, embedding is achieved with a set of software tools - Monte Carlo simulation software, as well as the tools for real data event reconstruction - accessed through a suite of scripts. These documents attempt to provide an introduction to the scripts and their use, but the STAR collaborator is referred to the separate web pages maintained for Simulations and Reconstruction for deeper questions of the processes involved in those respective tasks.
Please note, the general procedure for embedding is:
- The Physics Working Group Convenor submits an embedding request. The request will be assigned a request number and priority by the Analysis Coordinator, see this page for more information.
- The Embedding Coordinator will coordinate the distribution of embedding requests. Users who wish to run their own embedding are encouraged to do so - but it must be verified with the Embedding Coordinator.
- The Embedding Coordinator is responsible for the QA of all embedding requests. When the Embedding Coordinator has verified that the test sample produced is acceptable, full production is authorized.
For EC and ED(s) (OBSOLETE)
Embedding instructions for EC and ED(s)
Last updated on Sept/06/2011 by Christopher Powell
Current Embedding Coordinator (EC): Xianglei Zhu (zhux@rcf.rhic.bnl.gov)
Current Embedding Deputy (ED):
Current PDSF Point Of Contact (POC): Jeff Porter (RJPorter@lbl.gov)
-
Sept/06/2011: Update EC and ED
-
Nov/17/2010: Added some tickets for the records
-
Aug/02/2010: Update directory structure for LOG and Generator files
-
Jun/11/2010: Added approved chains by Lidia
-
May/29/2010: Modify procedures of 'Production chain options'
-
May/27/2010: Several minor fixes
-
May/25/2010: Added 'Production chain options', 'Locations of outputs etc at HPSS'
- May/24/2010: Update trigger id options
- May/21/2010: Update instructions for xml file, useful discussions
<!-- Generated by StRoot/macros/embedding/get_embedding_xml.pl on Mon Aug 2 15:26:13 PDT 2010 --> <?xml version="1.0" encoding="utf-8"?> <job maxFilesPerProcess="1" fileListSyntax="paths"> <command> <!-- Load library --> starver SL07e <!-- Set tags file directory --> setenv EMBEDTAGDIR /eliza3/starprod/tags/ppProductionJPsi/P06id <!-- Set year and day from filename --> setenv EMYEAR `StRoot/macros/embedding/getYearDayFromFile.pl -y ${FILEBASENAME}` setenv EMDAY `StRoot/macros/embedding/getYearDayFromFile.pl -d ${FILEBASENAME}` <!-- Set log files area --> setenv EMLOGS /project/projectdirs/star/embedding <!-- Set HPSS outputs/LOG path --> setenv EMHPSS /nersc/projects/starofl/embedding/ppProductionJPsi/JPsi_&FSET;_20100601/P06id.SL07e/${EMYEAR}/${EMDAY} <!-- Print out EMYEAR and EMDAY and EMLOGS --> echo EMYEAR : $EMYEAR echo EMDAY : $EMDAY echo EMLOGS : $EMLOGS echo EMHPSS : $EMHPSS <!-- Start job --> echo 'Executing bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); ...' root4star -b <<EOF std::vector<Int_t> triggers; triggers.push_back(117705); triggers.push_back(137705); triggers.push_back(117701); .L StRoot/macros/embedding/bfcMixer_TpcSvtSsd.C bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); .q EOF ls -la . cp $EMLOGS/P06id/LOG/$JOBID.log ${FILEBASENAME}.$JOBID.log cp $EMLOGS/P06id/LOG/$JOBID.elog ${FILEBASENAME}.$JOBID.elog <!-- New command to organize log files --> mkdir -p $EMLOGS/P06id/JPsi_20100601/LOG/&FSET; mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/ <!-- Archive in HPSS --> hsi "mkdir -p $EMHPSS; prompt; cd $EMHPSS; mput *.root; mput ${FILEBASENAME}.$JOBID.log; mput ${FILEBASENAME}.$JOBID.elog" </command> <!-- Define locations of log/elog files --> <stdout URL="file:/project/projectdirs/star/embedding/P06id/LOG/$JOBID.log"/> <stderr URL="file:/project/projectdirs/star/embedding/P06id/LOG/$JOBID.elog"/> <!-- Input daq files --> <input URL="file:/eliza3/starprod/daq/2006/st*"/> <!-- csh/list files --> <Generator> <Location>/project/projectdirs/star/embedding/P06id/JPsi_20100601/LIST</Location> </Generator> <!-- Put any locally-compiled stuffs into a sand-box --> <SandBox installer="ZIP"> <Package name="Localmakerlibs"> <File>file:./.sl44_gcc346/</File> <File>file:./StRoot/</File> <File>file:./pams/</File> </Package> </SandBox> </job>
1. Set up daq/tags files
<!-- Input daq files --> <input URL="file:/eliza3/starprod/daq/2006/st*"/>
<!-- Set tags file directory --> setenv EMBEDTAGDIR /eliza3/starprod/tags/ppProductionJPsi/P06id
> StRoot/macros/embedding/get_embedding_xml.pl -daq /eliza3/starprod/daq/2006 -tag /eliza3/starprod/tags/ppProductionJPsi/P06id
> StRoot/macros/embedding/get_embedding_xml.pl -tag /eliza3/starprod/tags/ppProductionJPsi/P06id -daq /eliza3/starprod/daq/2006
2. Running job, archive outputs into HPSS
Below is the descriptions to run the job (bfcMixer), save log files, put outputs/logs into HPSS.
<!-- Start job --> echo 'Executing bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); ...' root4star -b <<EOF std::vector<Int_t> triggers; triggers.push_back(117705); triggers.push_back(137705); triggers.push_back(117701); .L StRoot/macros/embedding/bfcMixer_TpcSvtSsd.C bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); .q EOF ls -la . cp $EMLOGS/P06id/LOG/$JOBID.log ${FILEBASENAME}.$JOBID.log cp $EMLOGS/P06id/LOG/$JOBID.elog ${FILEBASENAME}.$JOBID.elog <!-- New command to organize log files --> mkdir -p $EMLOGS/P06id/JPsi_20100601/LOG/&FSET; mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/ <!-- Archive in HPSS --> hsi "mkdir -p $EMHPSS; prompt; cd $EMHPSS; mput *.root; mput ${FILEBASENAME}.$JOBID.log; mput ${FILEBASENAME}.$JOBID.elog"
> StRoot/macros/embedding/get_embedding_xml.pl -mixer StRoot/macros/embedding/bfcMixer_Tpx.C
<= Run4 : bfcMixer_TpcOnly.C
Run5 - Run7 : bfcMixer_TpcSvtSsd.C
>= Run8 : bfcMixer_Tpx.C
> StRoot/macros/embedding/get_embedding_xml.pl -production P06id -lib SL07e -r 20100601 -trg ppProductionJPsi
<!-- Load library --> starver SL07e ... ... ls -la . cp $EMLOGS/P06id/LOG/$JOBID.log ${FILEBASENAME}.$JOBID.log cp $EMLOGS/P06id/LOG/$JOBID.elog ${FILEBASENAME}.$JOBID.elog <!-- New command to organize log files --> mkdir -p $EMLOGS/P06id/JPsi_20100601/LOG/&FSET; mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/ <!-- Archive in HPSS --> hsi "mkdir -p $EMHPSS; prompt; cd $EMHPSS; mput *.root; mput ${FILEBASENAME}.$JOBID.log; mput ${FILEBASENAME}.$JOBID.elog" ... ... <!-- csh/list files --> <Generator> <Location>/project/projectdirs/star/embedding/P06id/JPsi_20100601/LIST</Location> </Generator>
Error: No /project/projectdirs/star/embedding/P06id/JPsi_20100601/LIST exists. Stop. Make sure you've put the correct path for generator file.
mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/
3. Arguments in the bfcMixer

3-1. Particle id and particle name
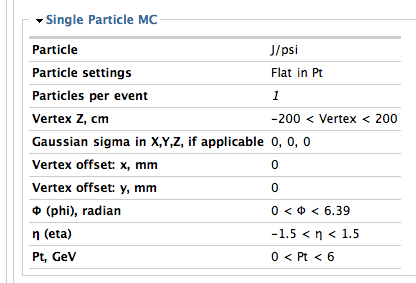
Particle Jpsi code=160 TrkTyp=4 mass=3.096 charge=0 tlife=7.48e-21,pdg=443 bratio= { 1, } mode= { 203, }
> StRoot/macros/embedding/get_embedding_xml.pl -geantid 160 -particle JPsi
3-2. Particle settings (how to simulate pt distribution)
> StRoot/macros/embedding/get_embedding_xml.pl -mode Strange
3-3. Multiplicity
> StRoot/macros/embedding/get_embedding_xml.pl -mult 0.05
3-4. Primary z-vertex range
> StRoot/macros/embedding/get_embedding_xml.pl -zmin -30.0 -zmax 30.0
3-5. Rapidity and transverse momentum range
> StRoot/macros/embedding/get_embedding_xml.pl -ymin -1.0 -ymax 1.0 -ptmin 0.0 -ptmax 6.0
3-6. Trigger id's
> StRoot/macros/embedding/get_embedding_xml.pl -trigger 117705
> StRoot/macros/embedding/get_embedding_xml.pl -trggier 117705 -trigger 137705 -trigger 117001
> StRoot/macros/embedding/get_embedding_xml.pl -trigger 117705 137705 ...
3-7. Production chain name
StRoot/macros/embedding/get_embedding_xml.pl -prodname P06idpp
> StRoot/macros/embedding/get_embedding_xml.pl -f -daq /eliza3/starprod/daq/2006 -tag /eliza3/starprod/tags/ppProductionJPsi/P06id \ -production P06id -lib SL07e -r 20100601 -trg ppProductionJPsi -geantid 160 -particle JPsi -ptmax 6.0 -trigger 117705 -trigger 137705 -trigger 117701 \ -prodname P06idpp
Production chain options
- ED will set up the proper bfcMixer macro with the correct chain options
-
EC and POC at PDSF will take a look and give feedback
-
Ask Lidia for her inputs, and verify the new chain with Lidia and Yuri
-
Enter all chains into Drupal embedding page for documentation
-
Commit bfcMixer into CVS
--------------------------------
P07ic CuCu production: TString prodP07icAuAu("P2005b DbV20070518 MakeEvent ITTF ToF ssddat spt SsdIt SvtIt pmdRaw OGridLeak OShortR OSpaceZ2 KeepSvtHit skip1row VFMCE -VFMinuit -hitfilt");
P08ic AuAu production: DbV20080418 B2007g ITTF adcOnly IAna KeepSvtHit VFMCE -hitfilt l3onl emcDY2 fpd ftpc trgd ZDCvtx svtIT ssdIT Corr5 -dstout
If spacecharge and gridleak corrections are on average instead of event by event then Corr5-> Corr4, OGridLeak3D, OSpaceZ2.
P08ie dAu production : DbV20090213 P2008 ITTF OSpaceZ2 OGridLeak3D beamLine, VFMCE TpcClu -VFMinuit -hitfilt
TString chain20pt("NoInput,PrepEmbed,gen_T,geomT,sim_T,trs,-ittf,-tpc_daq,nodefault);
P06id pp production : TString prodP06idpp("DbV20060729 pp2006b ITTF OSpaceZ2 OGridLeak3D VFMCE -VFPPVnoCTB -hitfilt");
P06ie pp production : TString prodP06iepp("DbV20060915 pp2006b ITTF OSpaceZ2 OGridLeak3D VFMCE -VFPPVnoCTB -hitfilt"); run# 7096005-7156040
TString prodP06iepp("DbV20061021 pp2006b ITTF OSpaceZ2 OGridLeak3D VFMCE -VFPPVnoCTB -hitfilt"); run# 7071001-709402
Locations of outputs, log files and back up of relevant codes at HPSS
/nersc/projects/starofl/embedding/${TRGSETUPNAME}/${PARTICLE}_&FSET;_${REAUESTID}/${PRODUCTION}.${LIBRARY}/${EMYEAR}/${EMDAY}
(starofl home) /home/starofl/embedding/CODE/${TRGSETUPNAME}/${PARTICLE}_${REQUESTID}/${PRODUCTION}.${LIBRARY}/${EMYEAR} (HPSS) /nersc/projects/starofl/embedding/CODE/${TRGSETUPNAME}/${PARTICLE}_${REQUESTID}/${PRODUCTION}.${LIBRARY}/${EMYEAR}
1. VFMCE chain option
2. Eta dip around eta ~ 0
3. EmbeddingShortCut chain option
will not apply corrections for simulated data (IdTruth > 0 && IdTruth
< 10000 && QA > 0.95).
Trs has to have it. TrsRS should not have it.
Yuri
Till this release this option was always "ON" by default.
The only need for back propagation is when you will use release >= SL10c
with Trs. This correction will be done in dev for nightly tests.
Yuri
4. Bug in StAssociationMaker
For EC and ED(s)
Embedding instructions for EC and ED(s)
Last updated on Oct/23/2018 by Xianglei Zhu
Current Embedding Coordinator (EC): Xianglei Zhu (zhux@tsinghua.edu.cn)
Current Embedding Deputy (ED): Derek Anderson (derekwigwam9@tamu.edu)
Current NERSC Point Of Contact (POC): Jeff Porter (RJPorter@lbl.gov) and Jan Balewski (balewski@lbl.gov)
- Jun/24/2018: initial version (copied from old instructions, still under construction)
- Instructions how to sample daq files and provide tags files
- Instructions how to set up xml file
- Production chain options
- Locations of outputs, log files and back up of relevant codes at HPSS
- Some useful discussions in hyper-news and bug-report
<!-- Generated by StRoot/macros/embedding/get_embedding_xml.pl on Mon Aug 2 15:26:13 PDT 2010 --> <?xml version="1.0" encoding="utf-8"?> <job maxFilesPerProcess="1" fileListSyntax="paths"> <command> <!-- Load library --> starver SL07e <!-- Set tags file directory --> setenv EMBEDTAGDIR /eliza3/starprod/tags/ppProductionJPsi/P06id <!-- Set year and day from filename --> setenv EMYEAR `StRoot/macros/embedding/getYearDayFromFile.pl -y ${FILEBASENAME}` setenv EMDAY `StRoot/macros/embedding/getYearDayFromFile.pl -d ${FILEBASENAME}` <!-- Set log files area --> setenv EMLOGS /project/projectdirs/star/embedding <!-- Set HPSS outputs/LOG path --> setenv EMHPSS /nersc/projects/starofl/embedding/ppProductionJPsi/JPsi_&FSET;_20100601/P06id.SL07e/${EMYEAR}/${EMDAY} <!-- Print out EMYEAR and EMDAY and EMLOGS --> echo EMYEAR : $EMYEAR echo EMDAY : $EMDAY echo EMLOGS : $EMLOGS echo EMHPSS : $EMHPSS <!-- Start job --> echo 'Executing bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); ...' root4star -b <<EOF std::vector<Int_t> triggers; triggers.push_back(117705); triggers.push_back(137705); triggers.push_back(117701); .L StRoot/macros/embedding/bfcMixer_TpcSvtSsd.C bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); .q EOF ls -la . cp $EMLOGS/P06id/LOG/$JOBID.log ${FILEBASENAME}.$JOBID.log cp $EMLOGS/P06id/LOG/$JOBID.elog ${FILEBASENAME}.$JOBID.elog <!-- New command to organize log files --> mkdir -p $EMLOGS/P06id/JPsi_20100601/LOG/&FSET; mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/ <!-- Archive in HPSS --> hsi "mkdir -p $EMHPSS; prompt; cd $EMHPSS; mput *.root; mput ${FILEBASENAME}.$JOBID.log; mput ${FILEBASENAME}.$JOBID.elog" </command> <!-- Define locations of log/elog files --> <stdout URL="file:/project/projectdirs/star/embedding/P06id/LOG/$JOBID.log"/> <stderr URL="file:/project/projectdirs/star/embedding/P06id/LOG/$JOBID.elog"/> <!-- Input daq files --> <input URL="file:/eliza3/starprod/daq/2006/st*"/> <!-- csh/list files --> <Generator> <Location>/project/projectdirs/star/embedding/P06id/JPsi_20100601/LIST</Location> </Generator> <!-- Put any locally-compiled stuffs into a sand-box --> <SandBox installer="ZIP"> <Package name="Localmakerlibs"> <File>file:./.sl44_gcc346/</File> <File>file:./StRoot/</File> <File>file:./pams/</File> </Package> </SandBox> </job>
1. Set up daq/tags files
<!-- Input daq files --> <input URL="file:/eliza3/starprod/daq/2006/st*"/>
<!-- Set tags file directory --> setenv EMBEDTAGDIR /eliza3/starprod/tags/ppProductionJPsi/P06id
> StRoot/macros/embedding/get_embedding_xml.pl -daq /eliza3/starprod/daq/2006 -tag /eliza3/starprod/tags/ppProductionJPsi/P06id
> StRoot/macros/embedding/get_embedding_xml.pl -tag /eliza3/starprod/tags/ppProductionJPsi/P06id -daq /eliza3/starprod/daq/2006
2. Running job, archive outputs into HPSS
Below is the descriptions to run the job (bfcMixer), save log files, put outputs/logs into HPSS.
<!-- Start job --> echo 'Executing bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); ...' root4star -b <<EOF std::vector<Int_t> triggers; triggers.push_back(117705); triggers.push_back(137705); triggers.push_back(117701); .L StRoot/macros/embedding/bfcMixer_TpcSvtSsd.C bfcMixer_TpcSvtSsd(1000, 1, 1, "$INPUTFILE0", "$EMBEDTAGDIR/${FILEBASENAME}.tags.root", 0, 6.0, -1.5, 1.5, -200, 200, 160, 1, triggers, "P08ic", "FlatPt"); .q EOF ls -la . cp $EMLOGS/P06id/LOG/$JOBID.log ${FILEBASENAME}.$JOBID.log cp $EMLOGS/P06id/LOG/$JOBID.elog ${FILEBASENAME}.$JOBID.elog <!-- New command to organize log files --> mkdir -p $EMLOGS/P06id/JPsi_20100601/LOG/&FSET; mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/ <!-- Archive in HPSS --> hsi "mkdir -p $EMHPSS; prompt; cd $EMHPSS; mput *.root; mput ${FILEBASENAME}.$JOBID.log; mput ${FILEBASENAME}.$JOBID.elog"
> StRoot/macros/embedding/get_embedding_xml.pl -mixer StRoot/macros/embedding/bfcMixer_Tpx.C
<= Run4 : bfcMixer_TpcOnly.C
Run5 - Run7 : bfcMixer_TpcSvtSsd.C
>= Run8 : bfcMixer_Tpx.C
> StRoot/macros/embedding/get_embedding_xml.pl -production P06id -lib SL07e -r 20100601 -trg ppProductionJPsi
<!-- Load library --> starver SL07e ... ... ls -la . cp $EMLOGS/P06id/LOG/$JOBID.log ${FILEBASENAME}.$JOBID.log cp $EMLOGS/P06id/LOG/$JOBID.elog ${FILEBASENAME}.$JOBID.elog <!-- New command to organize log files --> mkdir -p $EMLOGS/P06id/JPsi_20100601/LOG/&FSET; mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/ <!-- Archive in HPSS --> hsi "mkdir -p $EMHPSS; prompt; cd $EMHPSS; mput *.root; mput ${FILEBASENAME}.$JOBID.log; mput ${FILEBASENAME}.$JOBID.elog" ... ... <!-- csh/list files --> <Generator> <Location>/project/projectdirs/star/embedding/P06id/JPsi_20100601/LIST</Location> </Generator>
Error: No /project/projectdirs/star/embedding/P06id/JPsi_20100601/LIST exists. Stop. Make sure you've put the correct path for generator file.
mv $EMLOGS/P06id/LOG/$JOBID.* $EMLOGS/P06id/JPsi_20100601/LOG/&FSET;/
3. Arguments in the bfcMixer


3-1. Particle id and particle name
Particle Jpsi code=160 TrkTyp=4 mass=3.096 charge=0 tlife=7.48e-21,pdg=443 bratio= { 1, } mode= { 203, }
> StRoot/macros/embedding/get_embedding_xml.pl -geantid 160 -particle JPsi
3-2. Particle settings (how to simulate pt distribution)
> StRoot/macros/embedding/get_embedding_xml.pl -mode Strange
3-3. Multiplicity
> StRoot/macros/embedding/get_embedding_xml.pl -mult 0.05
3-4. Primary z-vertex range
> StRoot/macros/embedding/get_embedding_xml.pl -zmin -30.0 -zmax 30.0
3-5. Rapidity and transverse momentum range
> StRoot/macros/embedding/get_embedding_xml.pl -ymin -1.0 -ymax 1.0 -ptmin 0.0 -ptmax 6.0
3-6. Trigger id's
> StRoot/macros/embedding/get_embedding_xml.pl -trigger 117705
> StRoot/macros/embedding/get_embedding_xml.pl -trggier 117705 -trigger 137705 -trigger 117001
> StRoot/macros/embedding/get_embedding_xml.pl -trigger 117705 137705 ...
3-7. Production chain name
StRoot/macros/embedding/get_embedding_xml.pl -prodname P06idpp
> StRoot/macros/embedding/get_embedding_xml.pl -f -daq /eliza3/starprod/daq/2006 -tag /eliza3/starprod/tags/ppProductionJPsi/P06id \ -production P06id -lib SL07e -r 20100601 -trg ppProductionJPsi -geantid 160 -particle JPsi -ptmax 6.0 -trigger 117705 -trigger 137705 -trigger 117701 \ -prodname P06idpp
Production chain options
- ED will set up the proper bfcMixer macro with the correct chain options
- EC and POC at PDSF will take a look and give feedback
- Ask Lidia for her inputs, and verify the new chain with Lidia and Yuri
- Enter all chains into Drupal embedding page for documentation
- Commit bfcMixer into CVS
--------------------------------
P07ic CuCu production: TString prodP07icAuAu("P2005b DbV20070518 MakeEvent ITTF ToF ssddat spt SsdIt SvtIt pmdRaw OGridLeak OShortR OSpaceZ2 KeepSvtHit skip1row VFMCE -VFMinuit -hitfilt");
P08ic AuAu production: DbV20080418 B2007g ITTF adcOnly IAna KeepSvtHit VFMCE -hitfilt l3onl emcDY2 fpd ftpc trgd ZDCvtx svtIT ssdIT Corr5 -dstout
If spacecharge and gridleak corrections are on average instead of event by event then Corr5-> Corr4, OGridLeak3D, OSpaceZ2.
P08ie dAu production : DbV20090213 P2008 ITTF OSpaceZ2 OGridLeak3D beamLine, VFMCE TpcClu -VFMinuit -hitfilt
TString chain20pt("NoInput,PrepEmbed,gen_T,geomT,sim_T,trs,-ittf,-tpc_daq,nodefault);
P06id pp production : TString prodP06idpp("DbV20060729 pp2006b ITTF OSpaceZ2 OGridLeak3D VFMCE -VFPPVnoCTB -hitfilt");
P06ie pp production : TString prodP06iepp("DbV20060915 pp2006b ITTF OSpaceZ2 OGridLeak3D VFMCE -VFPPVnoCTB -hitfilt"); run# 7096005-7156040
TString prodP06iepp("DbV20061021 pp2006b ITTF OSpaceZ2 OGridLeak3D VFMCE -VFPPVnoCTB -hitfilt"); run# 7071001-709402
Locations of outputs, log files and back up of relevant codes at HPSS
/nersc/projects/starofl/embedding/${TRGSETUPNAME}/${PARTICLE}_&FSET;_${REAUESTID}/${PRODUCTION}.${LIBRARY}/${EMYEAR}/${EMDAY}
(starofl home) /home/starofl/embedding/CODE/${TRGSETUPNAME}/${PARTICLE}_${REQUESTID}/${PRODUCTION}.${LIBRARY}/${EMYEAR} (HPSS) /nersc/projects/starofl/embedding/CODE/${TRGSETUPNAME}/${PARTICLE}_${REQUESTID}/${PRODUCTION}.${LIBRARY}/${EMYEAR}
1. VFMCE chain option
2. Eta dip around eta ~ 0
3. EmbeddingShortCut chain option
will not apply corrections for simulated data (IdTruth > 0 && IdTruth
< 10000 && QA > 0.95).
Trs has to have it. TrsRS should not have it.
Yuri
Till this release this option was always "ON" by default.
The only need for back propagation is when you will use release >= SL10c
with Trs. This correction will be done in dev for nightly tests.
Yuri
4. Bug in StAssociationMaker
For Embedding Helpers (OBSOLETE)
Embedding instructions for Embedding Helpers
This instructions provide for embedding helpers how to prepare/submit the embedding jobs at PDSF
NOTE: This is specific instructions at PDSF, some procedures may not work at RCF
Current Embedding Coordinator (EC): Terence Tarnowsky (tarnowsk@nscl.msu.edu)
Christopher Powell (CBPowell@lbl.gov)
Current PDSF Point Of Contact (POC): Jeff Porter (RJPorter@lbl.gov)
-
Nov/8/2011: Updated EC and ED information to bring it in line with other webpages.
-
Apr/12/2011: Update Section 3.1, remove obsolete procedure for StPrepEmbedMaker
-
Feb/17/2011: Update Section 3.2
-
Nov/17/2010: Update to get afs token to access afs area
-
Sep/08/2010: Update memory limit in the batch at PDSF (Section 1)
-
Aug/25/2010: Update Section 3.2
-
May/25/2010: Update Section 3 (include klog, library and NOTE about VFMCE)
-
May/21/2010: Added "How to setup your environment at PDSF ?" and "Links"
-
May/20/2010: Updated instructions
Contents
1. How to set up your environment at PDSF ?
Please have a look at the "common issue: memory limit in batch"
and follow the procedure how to increase the memory limit in the batch jobs.
All EH should make this change before submitting any embedding production jobs.
2. Set up daq and tags files at PDSF
3. Production area setup
Below is the copy from the link above what you need to do in order to get afs token to access CVS
If so, you need to specify your afs account name explicitly as well.
> klog -cell rhic -principal YourRCFUserName
> cvs co StRoot/macros/embedding > cvs co StRoot/St_geant_Maker
> mv StRoot/St_geant_Maker/Embed/StPrepEmbedMaker* StRoot/St_geant_Maker/ > starver ${library} > cons
> cvs co pams/sim/gstar > cons
Please contact EC or ED whether bfcMixer (either bfcMixer_TpcSvtSsd.C or bfcMixer_Tpx.C) is ready to submit or not, and confirm which bfcMixer should be used for the current request.
4. Submit jobs
<!-- Put any locally-compiled stuffs into a sand-box --> <SandBox installer="ZIP"> <Package name="Localmakerlibs"> <File>file:./.sl44_gcc346/</File> <File>file:./StRoot/</File> <File>file:./pams/</File> </Package> </SandBox>
in your xml file. If you have anything other than above codes, please include them.
Please contact EC or ED if you are not clear enough which codes you need to include.
4-2. Submit jobs by scheduler
> star-submit-template -template embed_template.xml -entities FSET=200
4-3. Re-submitting jobs
Sometime you may need to modify something under "StRoot" or "pams", and recompile to fix some problems.
Each time you recompiled your local codes, you should clean up the current "Localmakerlibs.zip" and
"Localmakerlibs.package/" before starting resubmission. If you forgot to clean up the older "Localmakerlibs",
then the modification in the local codes will not reflect in the resubmitting jobs.
5. Back up macros, scripts, xml file and codes
6. Links
Obsolete documentations
Embedding Production Setup
This page describes how to set up embedding production. This procedure needs to be followed for any set of daq files/production version that requires embedding. Since this typically involves hacking the reconstruction chain, it is not advised that the typical STAR PA attempt this step. Please coordinate with a local embedding coordinator, and the overal Embedding coordinator (Olga).Note: The documentation here is very terse; it will be enriched as the documentation as a whole is iterated on. Patience is appreciated.
Get daq files from RCF.
| Grab a set of daq files from RCF which cover the lifetime of the run, the luminosity range experienced, and the conditions for the production. |
Rerun standard production but without corrections.
| bfc.C macros are located under ~starofl/bfc. Edit the submit.[Production] script to point to the daq files loaded (as above). |
Put tags files on disk.
| The results of the previous jobs will be .tags.root files located on HPSS. Retrieve the files, set a pointer for the tags files in the Production-specific directory under ~starofl/embedding. |
Now you're ready to start production.
Embedding Setup Off-site
Introduction
The purpose of this document is to describe step-by-step the setting up of embedding infrastructure on a remote site i.e. not at it's current home which is PDSF. It is based on the experience of setting up embedding at Birmingham's NP cluster (Bham). I will try to maintain a distinction between steps which are necessary in general and those which were specific to porting things to Bham. It should also be a useful guide for those wanting to run embedding at PDSF and needing to copy the relevant files into a suitable directory structure.Pre-requisites
Before trying to set up embedding on a remote site you should have:- a working local installation of the STAR library in which you are interested (or be satisified with your AFS-based library performance).
- a working mirror of the star database (or be satisfied with your connection to the BNL hosted db).
Collect scripts
The scripts are currently housed at PDSF in the 'starofl' account area. At the time of writing (and the time at which I set up embedding in Bham) they are not archived in CVS. The suggested way to collect them is to copy them into a directory in your own PDSF home account then tar and export it for installation on your local cluster. The top directory for embedding is /u/starofl/embedding . Under this directory there are several subdirectories of interest.- Those named after each production, e.g. P06ib which contain mixer macro and perl scripts
- Common which contains further subdirectories lists and csh and a submission perl script
- GSTAR which contains the kumac for running the simulation
mkdir embedding
cd embedding
mkdir Common
mkdir Common/lists
mkdir Common/csh
mkdir GSTAR
mkdir P06ib
mkdir P06ib/setup
Now it needs populating with the relevant files. In the following /u/user/embedding as an example of your new embedding directory in your user home directory.
cd /u/user/embedding
cp /u/starofl/embedding/getVerticesFromTags_v4.C .
cp -R /u/starofl/embedding/P06ib/EmbeddingLib_v4_noFTPC/ P06ib/
cp /u/starofl/embedding/P06ib/Embedding_sge_noFTPC.pl P06ib/
cp /u/starofl/embedding/P06ib/bfcMixer_v4_noFTPC.C P06ib/
cp /u/starofl/embedding/P06ib/submit.starofl.pl P06ib/submit.user.pl
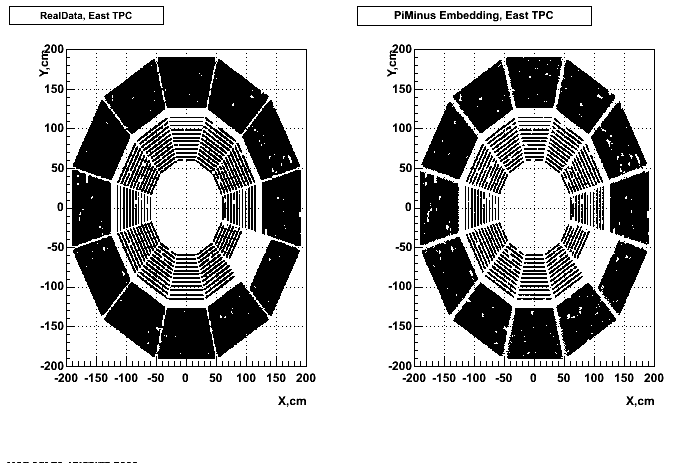
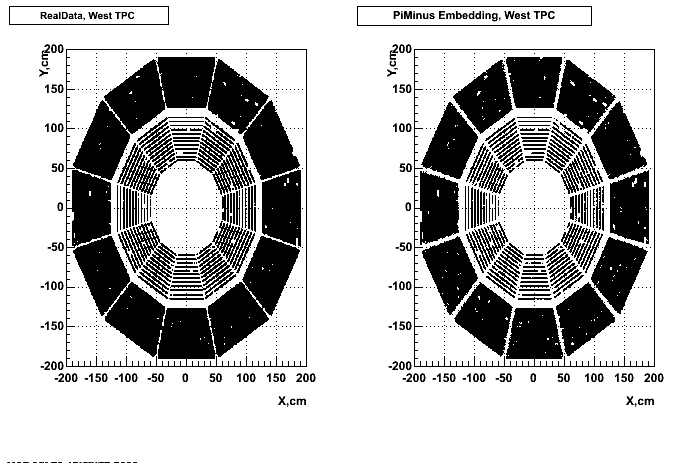
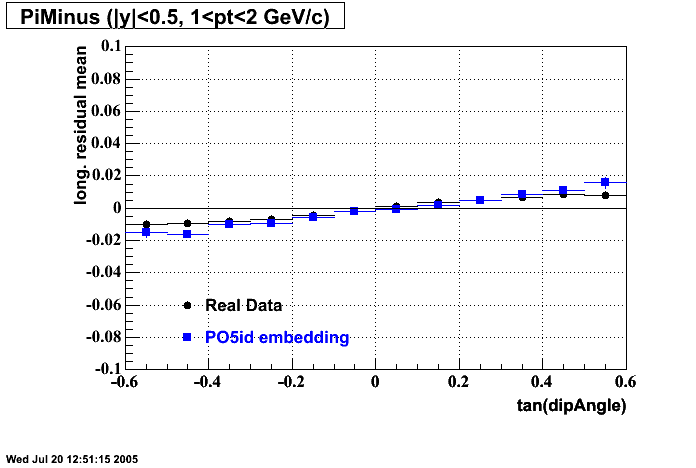
cp /u/starofl/embedding/P06ib/setup/Piminus_101_spectra.setup P06ib/setup/
cp /u/starofl/embedding/GSTAR/phasespace_P06ib_revfullfield.kumac GSTAR/
cp /u/starofl/embedding/GSTAR/phasespace_P06ib_fullfield.kumac GSTAR/
cp /u/starofl/embedding/Common/submit_sge.pl Common/
You now have all the files need to run embedding. There are further links to make but as you are going to export them to your own cluster you need to make the links afterwards.
Alternatively you can run embedding on PDSF from your home directory. There are a number of change to make first though because the various perl scripts have some paths relating to the starofl account inside them.
For those planning to export to a remote site you should tar and/or scp the data. I would recommend tar so that you can have the original package preserved in case something goes wrong. E.g.
tar -cvf embedding.tar embedding/
scp embedding.tar remoteuser@mycluster.blah.blah:/home/remoteuser
Obviously this step is unnecessary if you intend to run from your PDSF account although you may still want to create a tar file so that you can undo any changes which are wrong.
Login to your remote cluster and extract the archive. E.gcd /home/remoteuser
tar -xvf embedding.tar
Script changes
The most obvious thing you will find are a number of places inside the perl scripts where the path or location for other scripts appears in the code. These must be changed accordingly.
P06ib/Embedding_sge_noFTPC.pl
changes to e.g.changes to e.g.changes to e.g.changes to e.g.
P06ib/EmbeddingLib_v4_noFTPC/Process_object.pm
changes to e.g.
changes to e.g.
This is because the location of tcsh was different and probably will be for you too.
Common/submit_sge.pl
changes to e.g.
Change relates to parsing the name of the directory with daq files in to extract the 'data vault' and 'magnetic field' which form part of job name and are used by Embedding_sge_noFTPC.pl (This may not make much sense right now and needs the detailed docs on each component. It is actually just a way to pass a file list with the same basename as the job). In the original script the path to the data is something like/dante3/starprod/daq/2005/cuProductionMinBias/FullField
whereas on Bham cluster it is/star/data1/daq/2005/cuProductionMinBias/FullField
and thus the pattern match in perl has to change in order to extract the same information. If you have a choice then choose your directory names with care!changes to e.g.
Change relates to the line printing the job submission shell script that this perl script writes and submits. The first line had to be changed such that it can correctly be identified as a sh script. I am not sure how original can ever have worked?changes to e.g.
This line prints part of the job submission script where the options for the job are specified. In SGE the job options can be in the file and not just on the command line. The extra options for Bham relate to our SGE setup. The-q
option provides the name of the queue to use, otherwise it uses the default which I did not want in this case. The other extra options are to make the environment and working diretory correct as they were not the default for us. This is very specific to each cluster. If your cluster does not have SGE then I imagine extensive changes to the part writing the job submission script would be necessary. The scripts use the ability of SGE to have job arrays of similar jobs so you would have to emulate that somehow.
- getVerticesFromTags_v4.C - none
- GSTAR/phasespace_P06ib_fullfield.kumac, GSTAR/phasespace_P06ib_fullfield.kumac - actually there are changes but they only relate to redefining particle decay modes for (anti-)Ξ and (anti-)Ω to go 100% to charged modes of interest. This is only relevant for strangeness group
- P06ib/bfcMixer_v4_noFTPC.C - checked carefully that
chain3->SetFlagsline actually sets the same flags since Andrew and I had to change the same flags e.g. add GeantOut option after I made orginal copy - P06ib/EmbeddingLib_v4_noFTPC/Chain_object.pm - none
- P06ib/EmbeddingLib_v4_noFTPC/EmbeddingUtilities.pm - there are lines where you may have to add the run numbers of the daq files which you are using so that they are recognised as either full field or reversed full field. In this example (Cu+Cu embedding in P06ib) the lines begin
and. This is also something that Andrew and I both changed after I made the original copy. - P06ib/submit.user.pl - changes here relate to setup that you want to run and not to the cluster or directory you are using i.e. which setup file to use, what daq directories to use and any pattern match on the file names (usually for testing purposes to avoid filling the cluster with useless jobs) although you probably want to change the
line! - P06ib/setup/Piminus_101_spectra.setup - any changes here relate to the simulation parameters of the job that you want to do and not to the cluster or directory you are using
Create links
A number of links are required. For example in the /u/starofl/embedding/P06ib there are the following links:daq_dir_2005_cuPMBFF -> /dante3/starprod/daq/2005/cuProductionMinBias/FullFielddaq_dir_2005_cuPMBRFF -> /dante3/starprod/daq/2005/cuProductionMinBias/ReversedFullFielddaq_dir_2005_cuPMBHTFF -> /eliza5/starprod/daq/2005/cucuProductionHT/FullField/daq_dir_2005_cuPMBHTRFF -> /eliza5/starprod/daq/2005/cucuProductionHT/ReversedFullFieldtags_dir_cuHT_2005 -> /eliza5/starprod/embedding/tags/P06ibdata -> /eliza12/starprod/embedding/datalists ->../Common/listscsh-> ../Common/cshLOG-> ../Common/LOG
tags_dir_cu_2005 -> /dante3/starprod/tags/P06ib/2005
That is it! Some things will probably need to be adapted to your circumstances but it should give you a good idea of what to do
Author: Lee Barnby, University of Birmingham (using starembed account)
Modified: A. Rose, Lawrence Berkeley National Laboratory (using starembed account)
Modified Birmingham Files
Upload of modified embedding infrastructure files used on Birmingham NP cluster for Cu+Cu for (anti-)Λ and K0S embedding request.Production Management
1) Usually embedding jobs are run in "HPSS" mode so the files end up in HPSS (via FTP). To transfer them from HPSS to disk copy the perl script ~starofl/hjort/getEmbed.pl and modify as needed. This script does at least two things that are not possible with, e.g., a command line hsi command: it only gets the files needed (usually the .geant and .event files) and it changes the permissions after the transfers. Note that if you do the transfers shortly after running the jobs the files will probably still be on the HPSS disk cache and transfers will be much fast than getting the files from tapes.2) To clean up old embedding files make your own copy of ~starofl/hjort/embedAge.pl and use as needed. Note that $accThresh determines the maximum access time in days of files that will not be deleted.
Running Embedding
This page describes how to run embedding jobs once the daq files and tags files are in place (see other page about embedding production setup).Basics:
Embedding code is located in production specific directories: ~starofl/embedding/P0xxx. The basic job submission template is typically called submit.starofl.pl in that directory.Jobs are usually run by user starofl but personal accounts with group starprod membership will work, too (but test first as the group starprod write permissions typically are not in place by default).
The script to submit a set of jobs is submit.[user].pl. The script should be modified to submit an embedding set from the the configuration file
~starofl/embedding/[Production]/setup/[Particle]_[set]_[ID].setup
where
[Particle] is the particle type submitted (Piminus for GEANTID=9, as set inside file)
[set] is the file set submitted (more on this later)
[ID] is the embedding request numberTest procedure:
The best way to test a particular job configuration is to run a single job in "DISK" mode (by selecting a specific daq file in your submission). In this mode all of the intermediate files, scripts, logs, etc., are saved on disk. The location will be under the "data" link in the working directory. You can then go and figure out which script failed, hack as necessary and and try to make things work...Details:
QA Documentation
New embedding Base QA instructions
Current Embedding Coordinator (EC): Xianglei Zhu (zhux@tsinghua.edu.cn)
Current NERSC Point Of Contact (POC): Jeff Porter (RJPorter@lbl.gov) and Jan Balewski (balewski@lbl.gov)
-
Jan/9/2015: Update trigger selection on real data histogram making
-
Apr/19/2013: update responsible persons and add note about Omega mis-labeling issue
-
Apr/12/2011: Update NOTE in Section1
- Feb/17/2011: Update NOTE in Section1, and for checking out StAssociationMaker
-
Jan/27/2011: Update the Section 3 for some special decay daughters
-
Oct/25/2010: Added minimc reproduction in Section 1 and StAssociationMaker in Section
-
Jul/23/2010: Update the real data QA
-
May/21/2010: Minor style change, added "contents" and "Links" for "Embedding instructions for Embedding Helpers"
-
May/14/2010: Update rapidity/trigger selections for real data
- Apr/07/2010: Update the instructions on "drawEmbeddingQA.C"
- Feb/22/2010: Added how to change the z-vertex cut and about 'isEmbeddingOnly' flag in "drawEmbeddingQA.C"
- Jan/29/2010: Update the NOTE for "doEmbeddingQAMaker" (see below)
- Jan/22/2010: Update the documentation for the latest QA code
- Sep/21/2009: Add how to plot the QA histograms in the Section 3
- Sep/18/2009: Modify codes to "StEmbeddingQA*", and macro to "doEmbeddingQAMaker.C" (Please have a look at the instructions below carefully).
- Sep/17/2009: Modify Section 2.1
- Sep/15/2009: Add new embedding QA instructions
1. Produce minimc files
> cvs co StRoot/StMiniMcEvent > cvs co StRoot/StMiniMcMaker > cons
.sl53_gcc432/obj/StRoot/StMiniMcMaker/StMiniMcMaker.cxx: In member function 'void StMiniMcMaker::fillRcTrackInfo(StTinyRcTrack*, const StTrack*, const StTrack*, Int_t)': .sl53_gcc432/obj/StRoot/StMiniMcMaker/StMiniMcMaker.cxx:1622: error: 'const class StTrack' has no member named 'seedQuality'
1.1 Check out the relevant codes and macros in your working directory (suppose your working directory is ${work})
> cp /eliza8/rnc/hmasui/embedding/QA/StMiniHijing.C ${work}
- If the eliza8 is down, you can also copy the macro from the link below
- The current "StMiniHijing.C" has been slightly modified from the original version
to obtain a proper minimc filename based on the input geant.root file, see line 159-162 in "StMiniHijing.C"
159 TString filename = MainFile; 160 // int fileBeginIndex = filename.Index(filePrefix,0); 161 // filename.Remove(0,fileBeginIndex); 162 filename.Remove(0, filename.Last('/')+1);
- You don't need to modify the argument "filePrefix".
> cvs co StRoot/StAssociationMaker > cons
1.2 Run "StMiniHijing.C"
Either
> root4star -b -q StMiniHijing.C'(1000, "/eliza9/starprod/embedding/P08ie/dAu/Piplus_201_1233091546/Piplus_st_physics_adc_9020060_raw_2060010_201/st_physics_adc_9020060_raw_2060010.geant.root", "./")'
or
> root4star -b [0] .L StMiniHijing.C [1] StMiniHijing(1000, "/eliza9/starprod/embedding/P08ie/dAu/Piplus_201_1233091546/Piplus_st_physics_adc_9020060_raw_2060010_201/st_physics_adc_9020060_raw_2060010.geant.root", "./"); .... .... .... [2].q
- The 1st argument is maximum number of events.
- The 2nd argument is your input geant.root file.
- The 3rd argument is your output directory. For example, if you would like to put the output into
"./output" directory, you can set the 3rd argument as "./output/".
such as PiPlus_201_1233091546, PiPlus_202_1233091546 etc.
Since the filename of input geant.root's are usually identical among different groups,
you will overwrite your minimc.root if you put the minimc outputs under the same directory.
1.3 Make sure MC geantid is correct
> root4star st_physics_adc_9020060_raw_2060010.minimc.root [0] StMiniMcTree->Draw("mMcTracks.mGeantId")
[0] StMiniMcTree->Scan("mMcTracks.mGeantId") [1].q
2. Run QA codes
2.1 Check out the QA codes from CVS
QA macro
> cvs checkout StRoot/macros/embedding/doEmbeddingQAMaker.C
QA codes
> cvs checkout StRoot/StEmbeddingUtilities
2.2 Run the QA macro
2.2.1 QA for embedding outputs
and the output filename for your QA histograms is "embedding.root"
Either
> root4star -b -q doEmbeddingQAMaker.C'(2008, "P08ie", "minimc.list", "embedding.root")'
or
> root4star -b [0] .L doEmbeddingQAMaker.C [1] doEmbeddingQAMaker(2008, "P08ie", "minimc.list", "embedding.root"); ... ... ... [2] .q
The details of arguments can be found in the "doEmbeddingQAMaker.C"
- Output file name (4th argument, in this case "embedding.root") will be
automatically detemined according to the "year", "production"
and "particle name" if you leave it blank.
you can modify the 6th argument like
> roo4star -b[0] .L doEmbeddingQAMaker.C [1] doEmbeddingQAMaker(2008, "P08ie", "minimc.list", "", kTRUE, 60.0); ... ... ... [2] .q
where the 5th argument is the switch to analyze embedding (kTRUE) or real data (kFALSE).
2.2.2 QA for real data
> roo4star -b[0] .L doEmbeddingQAMaker.C [1] doEmbeddingQAMaker(2008, "P08ie", "mudst.list", "", kFALSE); ... ... ... [2] .q
2.2.3 Trigger selections and rapidity cuts for real data
- The trigger id can be also selected by
StEmbeddingQAUtilities::addTriggerIdCut(const UInt_t id)
StEmbeddingQAUtilities accept multiple trigger id's while current code assumes 1 trigger id per event,
The trigger id cut only affects the real data, not for the embedding outputs.
- You can also apply rapidity cut by
StEmbeddingQAUtilities::setRapidityCut(const Double_t ycut)
It would be good to have the same rapidity cut in the real data as the embedding production.
Please have a look at the simulation request page for rapidity cut or ask embedding helpers
what rapidity cuts they used for the productions.
3. Draw your QA results, compare with the real data
You can make the QA plots by "drawEmbeddingQA.C" under "StRoot/macros/embedding".
> cvs checkout StRoot/macros/embedding/drawEmbeddingQA.C
and "qa_real_2007_P08ic.root" (the output file format is automatically
determined like this if you leave the output filename blank in StEmbeddingQA).
> root4star -l drawEmbeddingQA.C'("./", "qa_embedding_2007_P08ic.root", "qa_real_2007_P08ic.root", 8, 10.0)'
First argument is the directory where the output PDF file is printed.
The default output directory is the current directory.
You can now check the QA histograms from embedding outputs only by
> root4star -l drawEmbeddingQA.C'("./", "qa_embedding_2007_P08ic.root", "qa_real_2007_P08ic.root", 8, 10.0, kTRUE)'
where the last argument 'isEmbeddingOnly' (default is kFALSE) is the switch
to draw the QA histograms for embedding outputs only if it is true.
If you name the output ROOT files by hand, you need to put the year and
production by yourself since those are used
for the output figure name and to print those informations in
the legend for each QA plot.
Suppose you have output ROOT files in your current directory: "qa_embedding.root" and "qa_real.root"
> root4star -l drawEmbeddingQA.C'("./", "qa_embedding.root", "qa_real.root", 2005, "P08ic", 8, 10.0, kFALSE)'
> root4star -l drawEmbeddingQA.C'("./", "qa_embedding_2007_P08ic.root", "qa_real_2007_P08ic.root", 8, 10, kFALSE, 37)'
maker->setParentGeantId(parentGeantId) ;
NOTE: Omegas are labelled as Ant-Omegas and vice versa. This is a known issue due to
of these libraries might fix it. For now, it has not been considered necessary to update all
embedding libraries to fix this issue. Please take note of this in all presentations regarding
(Anti-)Omega embedding.
4. Links
End of New embedding QA instructions
------------------------------------------------------------------------------------
This document is intended to describe the macros used during the quality assurance(Q/A) studies. This page is being updated today April 19 2009
* Macro : scan_embed_mc.C
After knowing the location of the minimc.root files use this macro to generate and output files with extension .root, in which all the histogramas for a basic QA had been filled. New histogramas had been added, ofr instacne a 3D histogram for Dca (pt, eta, dca) will give the distribution of dca as a function of pt and eta simultaneously. Same is done for the number of fit points (pr, eta, nfit). Also histograms to evaluate the randomness of the embedding files had been added to this macro.
* Macros: scan_embed_mudst.C
This macro hopefully you won't have to use it unless is requested. This macro is meant to generate and output root file with distributions coming from the MuDst (MuDst from Lidia) for a particular production. You will need just the location of the output file.
* Macro : plot_embed.C
This macro will take both outputs ( the one coming from minimc and that one coming from MuDst) and plot all the basic qa distributions for a particular production.
Hit Level
At the hit level, this is the documentation:* StEmbedHitsMaker.C
* doEmbedHits.C
* ScanHits.C
* PlotHits.C
See below the links for these macros
Scan-Hits.C
Plot_Hits.C
Plot_Dca.C
Plot_Nfit.C
Plot_embed
<code>//First run scan_embed.C to generate root file with all the histograms
// V. May 31 2007 - Cristina
#ifndef __CINT__
#include "TROOT.h"
#include "TSystem.h"
#include <iostream.h>
#include "TH1.h"
#include "TH2.h"
#include "TH3.h"
#include "TFile.h"
#include "TTree.h"
#include "TChain.h"
#include "TTreeHelper.h"
#include "TText.h"
#include "TLatex.h"
#include "TAttLine.h"
#include "TCanvas.h"
#endif
void plot_embed(Int_t id=9) {
gROOT->LoadMacro("~/macros/Utility.C"); //location of Utility.C
gStyle->SetOptStat(1);
//gStyle->SetOptTitle(0);
gStyle->SetOptDate(0);
gStyle->SetOptFit(0);
gStyle->SetPalette(1);
float mass2;
if (id == 8) { TString tag = "Piplus"; mass2 = 0.019;}
if (id == 9) { TString tag = "Piminus"; mass2 = 0.019;}
if (id == 11) { TString tag = "Kplus"; mass2 = 0.245;}
if (id == 12) { TString tag = "Kminus"; mass2 = 0.245;}
if (id == 14) { TString tag = "Proton"; mass2 = 0.880;}
if (id == 15) { TString tag = "Pbar"; mass2 = 0.880;}
if (id == 50) { TString tag = "Phi"; mass2 = 1.020;}
if (id == 2) { TString tag = "Eplus"; mass2 = 0.511;}
if (id == 1) { TString tag = "Dmeson"; mass2 = 1.864;}
char text1[80];
sprintf(text1,"P05_CuCu200_01_02_08");//this is going to show in all the histograms
char title[100],
char gif[100];
TString prod = "P05_CuCu200_01_02_08";
int nch1 = 0;
int nch2 = 1000;
/////////////////////////////////////////////////
//Cloning Histograms
/////////////////////////////////////////////////
f1 = new TFile ("~/data/P05_CuCu200_010208.root");
TH3D *hDca1 = (TH3D*)hDca -> Clone("hDca1");//DCA
TH3D *hNfit1 = (TH3D*)hNfit -> Clone("hNFit1");//Nfit
TH2D *hPtM_E1 = (TH2D*)hPtM_E -> Clone("hPtM_E1");//Energy Loss
TH2D *dedx1 = (TH2D*)dedx -> Clone("dedx1");
TH2D *dedxG1 = (TH2D*)dedxG -> Clone("dedxG1");
TH2D *vxy1 = (TH2D*)vxy -> Clone("vxy1");
TH1D *vz1 = (TH1D*)vz -> Clone("vz1");
TH1D *dvx1 = (TH1D*)dvx -> Clone("dvx1");
TH1D *dvy1 = (TH1D*)dvy -> Clone("dvy1");
TH1D *dvz1 = (TH1D*)dvz -> Clone("dvz1");
TH1D *PhiMc1 = (TH1D*)PhiMc -> Clone("PhiMc1");
TH1D *EtaMc1 = (TH1D*)EtaMc -> Clone("EtaMc1");
TH1D *PtMc1 = (TH1D*)PtMc -> Clone("PtMc1");
TH1D *PhiM1 = (TH1D*)PhiM -> Clone("PhiM1");
TH1D *EtaM1 = (TH1D*)EtaM -> Clone("EtaM1");
TH1D *PtM1 = (TH1D*)PtM -> Clone("PtM1");
TH2D *PtM_eff1 = (TH2D*)hPtM_eff ->Clone("PtM_eff1");//efficiency
//if you have MuDst hist
TH3D *hDca1r = (TH3D*)hDcaR -> Clone("hDca1r");
TH3D *hNfit1r = (TH3D*)hNfitR -> Clone("hNFit1r");
TH2D *dedx1R = (TH2D*)dedxR -> Clone("dedx1R");
/*
//use the following if you need to compare
f2 = new TFile ("~/data/test_P07ib_pi_10percent_10_03_07.root");
TH2D *PtM_eff2 = (TH2D*)hPtM_eff ->Clone("PtM_eff2");
TH3D *hNfit2 = (TH3D*)hNfit -> Clone("hNFit2");//Nfit
TH1D *PtM2 = (TH1D*)PtM -> Clone("PtM2");
TH1D *PtMc2 = (TH1D*)PtMc -> Clone("PtMc2");
f3 = new TFile ("~/data/test_P07ib_pi_10percent_10_12_07.root");
TH3D *hNfit3 = (TH3D*)hNfit -> Clone("hNFit3");//Nfit
TH2D *PtM_eff3 = (TH2D*)hPtM_eff ->Clone("PtM_eff3");
TH1D *PtM3 = (TH1D*)PtM -> Clone("PtM3");
TH1D *PtMc3 = (TH1D*)PtMc -> Clone("PtMc3");
*/
int nch1 = 0;
int nch2 = 1000;
Double_t pt[4]= {0.3,0.4, 0.5, 0.6};
////////////////////////////////////////////////////////////
//efficiency
/////////////////////////////////////////////////////////////
/*
TCanvas *c10= new TCanvas("c10","Efficiency",500, 500);
c10->SetGridx(0);
c10->SetGridy(0);
c10->SetLeftMargin(0.15);
c10->SetRightMargin(0.05);
//c10->SetTitleOffSet(0.1, "Y");
c10->cd;
PtM1->Rebin(2);
PtMc1->Rebin(2);
PtM1->Divide(PtMc1);
PtM1->SetLineColor(1);
PtM1->SetMarkerStyle(23);
PtM1->SetMarkerColor(1);
PtM1->Draw();
PtM1->SetXTitle ("pT (GeV/c)");
PtM1->SetAxisRange(0.0, 6.0, "X");
return;
/*
PtM2->Rebin(2);
PtMc2->Rebin(2);
PtM2->Divide(PtMc2);
PtM2->SetLineColor(9);
PtM2->SetMarkerStyle(21);
PtM2->SetMarkerColor(9);
PtM2->Draw("same");
PtM3->Rebin(2);
PtMc3->Rebin(2);
PtM3->Divide(PtMc2);
PtM3->SetLineColor(2);
PtM3->SetMarkerStyle(22);
PtM3->SetMarkerColor(2);
PtM3->Draw("same");
keySymbol(0.08, 1.0, text1, 1, 23, 0.04);
/////////////////////////////////////////////////////////////
//Vertex position
//////////////////////////////////////////////////////////
TCanvas *c6= new TCanvas("c6","Vertex position",600, 400);
c6->Divide(2,1);
c6_1->cd();
vz1->Rebin(2);
vz1->SetXTitle("Vertex Z");
vz1->Draw();
c6_2->cd();
vxy1->Draw("colz");
vxy1->SetAxisRange(-1.5, 1.5, "X");
vxy1->SetAxisRange(-1.5, 1.5, "Y");
vxy1->SetXTitle ("vertex X");
vxy1->SetYTitle ("vertex Y");
keyLine(.2, 1.05,text1,1);
c6->Update();
/////////////////////////////////////////////////////////////////////
//Dedx
////////////////////////////////////////////////////////////////////
TCanvas *c8= new TCanvas("c8","dEdx vs P",500, 500);
c8->SetGridx(0);
c8->SetGridy(0);
c8->SetLeftMargin(0.15);
c8->SetRightMargin(0.05);
c8->cd;
dedxG1->SetXTitle("Momentum P (GeV/c)");
dedxG1->SetYTitle("dE/dx");
dedxG1->SetAxisRange(0, 5., "X");
dedxG1->SetAxisRange(0, 8., "Y");
dedxG1->SetMarkerColor(1);
dedxG1->Draw();//"colz");
dedx1->SetMarkerStyle(7);
dedx1->SetMarkerSize(0.3);
dedx1->SetMarkerColor(2);
dedx1->Draw("same");
keyLine(.3, 0.87,"Embedded Tracks",2);
keyLine(.3, 0.82,"Ghost Tracks",1);
keyLine(.2, 1.05,text1,1);
c8->Update();
/////////////////////////////////////////////////////
//MIPS (just for pions)
/////////////////////////////////////////////////////
if (id==8 || id==9)
{
TCanvas *c9= new TCanvas("c9","MIPS",500, 500);
c9->SetGridx(0);
c9->SetGridy(0);
c9->SetLeftMargin(0.15);
c9->SetRightMargin(0.05);
c9->cd;
double pt1 = 0.4;
double pt2 = 0.6;
dedxG1 -> ProjectionX("rpx");
int blG = rpx->FindBin(pt1);
int bhG = rpx->FindBin(pt2);
cout<<blG<<endl;
cout<<bhG<<endl;
dedxG1->ProjectionY("rpy",blG,bhG);
rpy->SetTitle("MIPS");
rpy->SetMarkerStyle(22);
// rpy->SetMarkerColor(2);
rpy->SetAxisRange(1.3, 4, "X");
//dedxG1->Draw();
dedx1->ProjectionX("mpx");
int blm = mpx->FindBin(pt1);
int bhm = mpx->FindBin(pt2);
cout<<blm<<endl;
cout<<bhm<<endl;
dedx1->ProjectionY("mpy", blm,bhm);
mpy->SetAxisRange(0.5, 6, "X");
mpy->SetMarkerStyle(22);
mpy->SetMarkerColor(2);
float max_rpy = rpy->GetMaximum();
max_rpy /= 1.*mpy->GetMaximum();
mpy->Scale(max_rpy);
cout<<"max_rpy is: "<<max_rpy<<endl;
cout<<"mpy is: "<<mpy<<endl;
rpy->Sumw2();
mpy->Sumw2();
rpy->Fit("gaus","","",1,4);
mpy->Fit("gaus","","", 1, 4);
mpy->GetFunction("gaus")->SetLineColor(2);
rpy->SetAxisRange(0.5 ,6.0, "X");
mpy->Draw();
rpy->Draw("same");
float mipMc = mpy->GetFunction("gaus")->GetParameter(1);//mean
float mipGhost = rpy->GetFunction("gaus")->GetParameter(1);
float sigmaMc = mpy->GetFunction("gaus")->GetParameter(2);//mean
float sigmaGhost = rpy->GetFunction("gaus")->GetParameter(2);
char label1[80];
char label2[80];
char label3[80];
char label4[80];
sprintf(label1,"mip MC %.3f",mipMc);
sprintf(label2,"mip Ghost %.3f",mipGhost);
sprintf(label3,"sigma MC %.3f",sigmaMc);
sprintf(label4,"sigma Ghost %.3f",sigmaGhost);
keySymbol(.5, .9, label1,2,1);
keySymbol(.5, .85, label3,2,1);
keySymbol(.5, .75, label2,1,1);
keySymbol(.5, .70, label4,1,1);
keyLine(.2, 1.05,text1,1);
char name[30];
sprintf(name,"%.2f GeV/c < Pt < %.2f GeV/c",pt1, pt2);
keySymbol(0.3, 0.65, name, 1, 1, 0.04);
c9->Update();
}//close if pion
/////////////////////////////////////////////////////////////////////////
//Energy loss
//////////////////////////////////////////////////////////////////////////
TCanvas *c7= new TCanvas("c7","Energy Loss",400, 400);
c7->SetGridx(0);
c7->SetGridy(0);
c7->SetLeftMargin(0.20);
c7->SetRightMargin(0.05);
c7->cd;
hPtM_E->ProfileX("pfx");
pfx->SetAxisRange(-0.01, 0.01, "Y");
pfx->SetAxisRange(0, 6, "X");
pfx->GetYaxis()->SetDecimals();
pfx->SetMarkerStyle(23);
pfx->SetMarkerSize(0.038);
pfx->SetMarkerColor(4);
pfx->SetLineColor(4);
pfx->SetXTitle ("Pt-Reco");
pfx->SetYTitle ("ptM - PtMc");
pfx->SetTitleOffset(2,"Y");
pfx->Draw();
/*hPtM_E1->ProfileX("pfx1");
pfx1->SetAxisRange(-0.007, 0.007, "Y");
pfx1->GetYaxis()->SetDecimals();
pfx1->SetLineColor(2);
pfx1->SetMarkerStyle(21);
pfx1->SetMarkerSize(0.035);
pfx1->SetXTitle ("Pt-Reco");
pfx1->SetYTitle ("ptM - PtMc");
pfx1->SetTitleOffset(2,"Y");
pfx1->Draw("same");
c7->Update();
//////////////////////////////////////////////////////
//pt
//////////////////////////////////////////////////////
TCanvas *c2= new TCanvas("c2","pt",500, 500);
c2->SetGridx(0);
c2->SetGridy(0);
c2->SetTitle(0);
c2->cd();
//embedded
PtMc1->Rebin(2);
PtMc1->SetLineColor(2);
PtMc1->SetMarkerStyle(20);
PtMc1->SetMarkerColor(2);
PtMc1->Draw();
PtMc1->SetXTitle ("pT (GeV/c)");
PtMc1->SetAxisRange(0.0, 6.0, "X");
//Reco
PtM1->Rebin(2);
PtM1->SetMarkerStyle(20);
PtM1->SetMarkerColor(1);
PtM1->Draw("same");
keySymbol(.2, 1.05,text1,1);
keyLine(.3, 0.20,"Embeded-McTracks",2);
keyLine(.3, 0.16,"Matched Pairs",1);
// keyLine(.4, 0.82,"Previous Embedding",4);
c2->Update();
//////////////////////////////////////////////////////////////////
//phi
/////////////////////////////////////////////////////////////////
TCanvas *c5= new TCanvas("c5","pt",500, 500);
//c5->Divide(2,1);
c5->SetGridx(0);
c5->SetGridy(0);
c5->SetTitle(0);
c5->cd();
//embedded
PhiMc1->Rebin(2);
PhiMc1->SetLineColor(2);
PhiMc1->SetMarkerStyle(20);
PhiMc1->SetMarkerColor(2);
PhiMc1->Draw();
PhiMc1->SetXTitle ("Phi");
PhiMc1->SetAxisRange(-4, 4.0, "X");
//Reco
PhiM1->Rebin(2);
PhiM1->SetMarkerStyle(20);
PhiM1->SetMarkerColor(1);
PhiM1->Draw("same");
//Previous
// PhiM ->Rebin(2);
// PtM->SetLineColor(4);
// PtM->SetMarkerColor(4);
//PtM->Draw("same");
TLatex l;
l.DrawLatex(7.0, 450.0, prod);
keySymbol(.2, 1.05,text1,1);
keyLine(.3, 0.20,"Embeded-McTracks",2);
keyLine(.3, 0.16,"Reco - Matched Pairs",1);
// keyLine(.4, 0.82,"Previous Embedding",4);
c2->Update();
c5->Update();
/////////////////////////////////////
//eta
///////////////////////////////////////////////////////////////
TCanvas *c2= new TCanvas("c2","Eta",500, 500);
c2->SetGridx(0);
c2->SetGridy(0);
c2->SetTitle(0);
c2->cd();
//embedded
EtaMc1->Rebin(2);
EtaMc1->SetLineColor(2);
EtaMc1->SetMarkerStyle(20);
EtaMc1->SetMarkerColor(2);
EtaMc1->Draw();
EtaMc1->SetXTitle ("Eta");
EtaMc1->SetAxisRange(-1.2, 1.2, "X");
//Reco
EtaM1->Rebin(2);
EtaM1->SetMarkerStyle(20);
EtaM1->SetMarkerColor(1);
EtaM1->Draw("same");
TLatex l;
l.DrawLatex(7.0, 450.0, prod);
keySymbol(.2, 1.05,text1,1);
keyLine(.3, 0.20,"Embeded-McTracks",2);
keyLine(.3, 0.16,"Reco - Matched Pairs",1);
// keyLine(.4, 0.82,"Previous Embedding",4);
c2->Update();
/////////////////////////////////////////////////////////////
//DCA
////////////////////////////////////////////////////////////
TCanvas *c= new TCanvas("c","DCA",800, 400);
c->Divide(3,1);
c->SetGridx(0);
c->SetGridy(0);
//Matched (Bins for Multiplicity)
TH1D *hX1 = (TH1D*)hDca1->Project3D("X");
Int_t bin_nch1 = hX1->FindBin(nch1);
Int_t bin_nch2 = hX1->FindBin(nch2);//this should be the same for both graphs (for 3 graphs)
//Bins for Pt
TString name1 = "hDca1";
TString namer1 = "hDcar1";
TString name = "hDca";
TH1D *hY1 = (TH1D*)hDca1->Project3D("Y");
TH1D *hY1_r = (TH1D*)hDca1r->Project3D("Y");
TH1D *hY = (TH1D*)hDca->Project3D("Y");
Double_t Sum1_MC;
Double_t Sum1_Real;
Double_t Sum_MC;
for(Int_t i=0; i<3 ; i++)
{
c->cd(i+1);
Int_t bin_ptl_1 = hY1->FindBin(pt[i]);
Int_t bin_pth_1 = hY1->FindBin(pt[i+1]);
TH1D *hDcaNew1= (TH1D*)hDca1->ProjectionZ(name1+i,bin_nch1, bin_nch2, bin_ptl_1, bin_pth_1);
Sum1_MC = hDcaNew1 ->GetSum();
cout<<Sum1_MC<<endl;
hDcaNew1->Scale(1./Sum1_MC);
hDcaNew1 ->SetLineColor(2);
hDcaNew1->Draw();
hDcaNew1->SetXTitle("Dca (cm)");
sprintf(title," %.2f GeV < pT < %.2f GeV, %d < nch < %d", pt[i],pt[i+1],nch1,nch2);
hDcaNew1->SetTitle(title);
//----Now MuDSt
Int_t bin_ptrl_1r = hY1_r->FindBin(pt[i]);
Int_t bin_ptrh_1r = hY1_r->FindBin(pt[i+1]);
TH1D *hDca_r1= (TH1D*)hDca1r->ProjectionZ(namer1+i,bin_nch1, bin_nch2, bin_ptrl_1r, bin_ptrh_1r);
Sum1_Real = hDca_r1 ->GetSum();
cout<<Sum1_Real<<endl;
hDca_r1->Scale(1./Sum1_Real);
hDca_r1->Draw("same");
//Now Previous Embedding
Int_t bin_ptl = hY->FindBin(pt[i]);
Int_t bin_pth = hY->FindBin(pt[i+1]);
TH1D *hDcaNew = (TH1D*)hDca->ProjectionZ(name+i,bin_nch1, bin_nch2, bin_ptl, bin_pth);
Sum_MC = hDcaNew ->GetSum();
cout<<Sum_MC<<endl;
hDcaNew->Scale(1./Sum_MC);
hDcaNew ->SetLineColor(4);
hDcaNew->Draw("same");
keySymbol(.4, .95,text1,1,1);
keyLine(0.4, 0.90,"MC- Matched Pairs",2);
keyLine(0.4, 0.85,"MuDst",1);
keyLine(0.4, 0.80,"Previous Embedding P06ib",4);
}
c->Update();
///////////////////////////////////////////////////
//NFIT
////////////////////////////////////////////////////
TCanvas *c1= new TCanvas("c1","NFIT",800, 400);
c1->Divide(3,1);
c1->SetGridx(0);
c1->SetGridy(0);
//Bins for Multiplicity -Matched tracks
TH1D *hX1 = (TH1D*)hNfit1->Project3D("X");
Int_t bin_nch1 = hX1->FindBin(nch1);
Int_t bin_nch2 = hX1->FindBin(nch2);//this should be the same for both graphs (for 3 graphs)
//Bins for Pt
TString name_nfit1 = "hNfit1";
TString name_nfitr1 = "hNfitr1";
TString name_nfit = "hNfit";
TH1D *hY1 = (TH1D*)hNfit1->Project3D("Y");
TH1D *hY1_r = (TH1D*)hNfit1r->Project3D("Y");
TH1D *hY = (TH1D*)hNfit->Project3D("Y");
Double_t Sum1_Nfit_MC;
Double_t Sum1_Nfit_Real;
Double_t Sum__Nfit_MC;
for(Int_t i=0; i<3 ; i++)
{
c1->cd(i+1);
Int_t bin_ptl_1 = hY1->FindBin(pt[i]);
Int_t bin_pth_1 = hY1->FindBin(pt[i+1]);
TH1D *hNfitNew1= (TH1D*)hNfit1->ProjectionZ(name_nfit1+i,bin_nch1, bin_nch2, bin_ptl_1, bin_pth_1);
Sum1_Nfit_MC = hNfitNew1 ->GetSum();
cout<<Sum1_Nfit_MC<<endl;
hNfitNew1->Scale(1./Sum1_Nfit_MC);
hNfitNew1 ->SetLineColor(2);
hNfitNew1->Draw();
hNfitNew1->SetXTitle("Nfit");
sprintf(title," %.2f GeV < pT < %.2f GeV, %d < nch < %d", pt[i],pt[i+1],nch1,nch2);
hNfitNew1->SetTitle(title);
//----Now MuDSt
Int_t bin_ptrl_1r = hY1_r->FindBin(pt[i]);
Int_t bin_ptrh_1r = hY1_r->FindBin(pt[i+1]);
TH1D *hNfit_r1= (TH1D*)hNfit1r->ProjectionZ(name_nfitr1+i,bin_nch1, bin_nch2, bin_ptrl_1r, bin_ptrh_1r);
Sum1_Nfit_Real = hNfit_r1 ->GetSum();
cout<<Sum1_Nfit_Real<<endl;
hNfit_r1->Scale(1./Sum1_Nfit_Real);
hNfit_r1->Draw("same");
//Now Previous Embedding
Int_t bin_ptl = hY->FindBin(pt[i]);
Int_t bin_pth = hY->FindBin(pt[i+1]);
TH1D *hNfitNew = (TH1D*)hNfit->ProjectionZ(name_nfit+i,bin_nch1, bin_nch2, bin_ptl, bin_pth);
Sum_Nfit_MC = hNfitNew ->GetSum();
cout<<Sum__Nfit_MC<<endl;
hNfitNew->Scale(1./Sum_Nfit_MC);
hNfitNew ->SetLineColor(4);
hNfitNew->Draw("same");
///*T = new TBox(40, 0, 50, 0.01);
//T->SetLineColor(2);
//T->SetLineWidth(2);
//T->Draw("same");
//check this....
keySymbol(.2, .95,text1,1,1);
keyLine(0.2, 0.90,"MC- Matched Pairs",2);
keyLine(0.2, 0.85,"MuDst",1);
keyLine(0.2, 0.80,"Previous Embedding P06ib",4);
}
return;
}
</code>
scan_embed.C
Notes
Overview
The document is intended as a forum for embedding group members to share information and notes.
Jobs Done
P06ib:6050022_1050001.28606: /eliza5/starprod/embedding/P06ib
6050022_1050001.7247: /eliza5/starprod/embedding/P06ib
6050022_1060001.3717: /eliza5/starprod/embedding/P06ib
6050022_1070001.10533: /eliza5/starprod/embedding/P06ib
6050022_1070001.15557: /eliza5/starprod/embedding/P06ib
6050022_1080001.26181: /eliza5/starprod/embedding/P06ib
6050022_1080002.25705: /eliza5/starprod/embedding/P06ib
6050022_1080002.6089: /eliza5/starprod/embedding/P06ib
6050022_2050001.28441: /eliza5/starprod/embedding/P06ib
6050022_2060001.20921: /eliza5/starprod/embedding/P06ib
6050022_2060001.2233: /eliza5/starprod/embedding/P06ib
6050022_2060002.29805: /eliza5/starprod/embedding/P06ib
6050022_2060002.31396: /eliza5/starprod/embedding/P06ib
6050022_2070001.1384: /eliza5/starprod/embedding/P06ib
6050022_2070001.27931: /eliza5/starprod/embedding/P06ib
6050022_3050001.26648: /eliza5/starprod/embedding/P06ib
6050022_3050002.20435: /eliza5/starprod/embedding/P06ib
6050022_3060001.32622: /eliza5/starprod/embedding/P06ib
6050022_3060001.6911: /eliza5/starprod/embedding/P06ib
6050022_3060002.10366: /eliza5/starprod/embedding/P06ib
6050022_3060002.5583: /eliza5/starprod/embedding/P06ib
6052072_1050001.15271: /eliza5/starprod/embedding/P06ib
6052072_1060001.18062: /eliza5/starprod/embedding/P06ib
6052072_1080001.20338: /eliza5/starprod/embedding/P06ib
6052072_1080002.27976: /eliza5/starprod/embedding/P06ib
6052072_2070001.27969: /eliza5/starprod/embedding/P06ib
6052072_2070002.21296: /eliza5/starprod/embedding/P06ib
6052072_2080001.17830: /eliza5/starprod/embedding/P06ib
6052072_3070001.15967: /eliza5/starprod/embedding/P06ib
AOmega_111_strange: /eliza5/starprod/embedding/P06ib
AOmega_112_strange: /eliza5/starprod/embedding/P06ib
AOmega_121_strange: /eliza5/starprod/embedding/P06ib
AOmega_122_strange: /eliza5/starprod/embedding/P06ib
AOmega_123_strange: /eliza5/starprod/embedding/P06ib
AXi_101_strange: /eliza5/starprod/embedding/P06ib
AXi_102_strange: /eliza5/starprod/embedding/P06ib
AXi_111_strange: /eliza5/starprod/embedding/P06ib
AXi_112_strange: /eliza5/starprod/embedding/P06ib
AXi_113_strange: /eliza5/starprod/embedding/P06ib
E_101_1154003879: /eliza5/starprod/embedding/P06ib
E_102_1154003879: /eliza5/starprod/embedding/P06ib
E_106_1154003879: /eliza5/starprod/embedding/P06ib
E_107_1154003879: /eliza5/starprod/embedding/P06ib
E_201_1154003879: /eliza5/starprod/embedding/P06ib
E_202_1154003879: /eliza5/starprod/embedding/P06ib
E_203_1154003879: /eliza5/starprod/embedding/P06ib
E_204_1154003879: /eliza5/starprod/embedding/P06ib
E_205_1154003879: /eliza5/starprod/embedding/P06ib
E_206_1154003879: /eliza5/starprod/embedding/P06ib
E_207_1154003879: /eliza5/starprod/embedding/P06ib
Eplus_502_1154003879: /eliza5/starprod/embedding/P06ib
Lambda_201_strange: /eliza5/starprod/embedding/P06ib
Omega_111_strange: /eliza5/starprod/embedding/P06ib
Omega_112_strange: /eliza5/starprod/embedding/P06ib
Omega_113_strange: /eliza5/starprod/embedding/P06ib
Phi_101_1163628205: /eliza5/starprod/embedding/P06ib
Pi0_101_1163628205: /eliza5/starprod/embedding/P06ib
Pi0_102_1163628205: /eliza5/starprod/embedding/P06ib
Pi0_103_1163628205: /eliza5/starprod/embedding/P06ib
Xi_111_strange: /eliza5/starprod/embedding/P06ib
Xi_112_strange: /eliza5/starprod/embedding/P06ib
Xi_113_strange: /eliza5/starprod/embedding/P06ib
P05if:
Eminus_503_1154003879: /eliza12/starprod/embedding/P05if
Eminus_504_1154003879: /eliza12/starprod/embedding/P05if
Eplus_503_1154003879: /eliza12/starprod/embedding/P05if
Eplus_504_1154003879: /eliza12/starprod/embedding/P05if
Eplus_505_1154003879: /eliza12/starprod/embedding/P05if
P05ic:
K0Short_122_strange: /auto/pdsfdv40/starprod/embedding/P05ic
K0Short_124_strange: /auto/pdsfdv40/starprod/embedding/P05ic
K0Short_131_strange: /auto/pdsfdv40/starprod/embedding/P05ic
K0Short_132_strange: /auto/pdsfdv40/starprod/embedding/P05ic
Lambda_112_strange: /auto/pdsfdv40/starprod/embedding/P05ic
Lambda_114_strange: /auto/pdsfdv40/starprod/embedding/P05ic
Lambda_122_strange: /auto/pdsfdv40/starprod/embedding/P05ic
Lambda_124_strange: /auto/pdsfdv40/starprod/embedding/P05ic
Lambda_131_strange: /auto/pdsfdv40/starprod/embedding/P05ic
Lambda_132_strange: /auto/pdsfdv40/starprod/embedding/P05ic
Piminus_101_spectra: /auto/pdsfdv40/starprod/embedding/P05ic
Piplus_101_spectra: /auto/pdsfdv40/starprod/embedding/P05ic
Pbar_201_spectra: /auto/pdsfdv45/starprod/embedding/P05ic
Pbar_202_spectra: /auto/pdsfdv45/starprod/embedding/P05ic
Phi_115_1118150698: /auto/pdsfdv45/starprod/embedding/P05ic
Phi_116_1118150698: /auto/pdsfdv45/starprod/embedding/P05ic
Phi_117_1118150698: /auto/pdsfdv45/starprod/embedding/P05ic
Piplus_201_spectra: /auto/pdsfdv45/starprod/embedding/P05ic
Piplus_202_spectra: /auto/pdsfdv45/starprod/embedding/P05ic
K0Short_101_strange: /dante3/starprod/embedding/P05ic
K0Short_102_strange: /dante3/starprod/embedding/P05ic
Jpsi_102_spectra: /eliza1/starprod/embedding/P05ic
Jpsi_103_spectra: /eliza1/starprod/embedding/P05ic
Jpsi_104_spectra: /eliza1/starprod/embedding/P05ic
Jpsi_105_spectra: /eliza1/starprod/embedding/P05ic
Piplus_201_ftpcw: /eliza1/starprod/embedding/P05ic
Sigma1385minus_301_strange: /eliza1/starprod/embedding/P05ic
Sigma1385plus_301_strange: /eliza1/starprod/embedding/P05ic
Sigma1385plus_302_strange: /eliza1/starprod/embedding/P05ic
Omega_101_strange: /eliza5/starprod/embedding/P05ic
Omega_102_strange: /eliza5/starprod/embedding/P05ic
Photon_201_spectra: /eliza5/starprod/embedding/P05ic
Photon_202_spectra: /eliza5/starprod/embedding/P05ic
Photon_203_spectra: /eliza5/starprod/embedding/P05ic
Photon_204_spectra: /eliza5/starprod/embedding/P05ic
Photon_205_spectra: /eliza5/starprod/embedding/P05ic
Photon_206_spectra: /eliza5/starprod/embedding/P05ic
Photon_207_spectra: /eliza5/starprod/embedding/P05ic
Photon_208_spectra: /eliza5/starprod/embedding/P05ic
Photon_209_spectra: /eliza5/starprod/embedding/P05ic
Photon_210_spectra: /eliza5/starprod/embedding/P05ic
Photon_211_spectra: /eliza5/starprod/embedding/P05ic
Photon_212_spectra: /eliza5/starprod/embedding/P05ic
Photon_213_spectra: /eliza5/starprod/embedding/P05ic
Photon_214_spectra: /eliza5/starprod/embedding/P05ic
Photon_215_spectra: /eliza5/starprod/embedding/P05ic
Photon_301_spectra: /eliza5/starprod/embedding/P05ic
Photon_302_spectra: /eliza5/starprod/embedding/P05ic
Photon_303_spectra: /eliza5/starprod/embedding/P05ic
Photon_304_spectra: /eliza5/starprod/embedding/P05ic
Photon_305_spectra: /eliza5/starprod/embedding/P05ic
Photon_306_spectra: /eliza5/starprod/embedding/P05ic
Photon_308_spectra: /eliza5/starprod/embedding/P05ic
Photon_309_spectra: /eliza5/starprod/embedding/P05ic
Photon_310_spectra: /eliza5/starprod/embedding/P05ic
Photon_311_spectra: /eliza5/starprod/embedding/P05ic
Photon_312_spectra: /eliza5/starprod/embedding/P05ic
Photon_313_spectra: /eliza5/starprod/embedding/P05ic
Photon_314_spectra: /eliza5/starprod/embedding/P05ic
Photon_315_spectra: /eliza5/starprod/embedding/P05ic
Photon_501_spectra: /eliza5/starprod/embedding/P05ic
Photon_502_spectra: /eliza5/starprod/embedding/P05ic
Photon_503_spectra: /eliza5/starprod/embedding/P05ic
Photon_504_spectra: /eliza5/starprod/embedding/P05ic
Piminus_101_spectra: /eliza5/starprod/embedding/P05ic
Piminus_118_spectra: /eliza5/starprod/embedding/P05ic
Piminus_119_spectra: /eliza5/starprod/embedding/P05ic
Piminus_120_spectra: /eliza5/starprod/embedding/P05ic
Piminus_121_spectra: /eliza5/starprod/embedding/P05ic
Piminus_122_spectra: /eliza5/starprod/embedding/P05ic
Piplus_118_spectra: /eliza5/starprod/embedding/P05ic
Piplus_119_spectra: /eliza5/starprod/embedding/P05ic
Piplus_120_spectra: /eliza5/starprod/embedding/P05ic
Piplus_121_spectra: /eliza5/starprod/embedding/P05ic
Piplus_122_spectra: /eliza5/starprod/embedding/P05ic
Piplus_212_ftpcw: /eliza5/starprod/embedding/P05ic
Xi_101_strange: /eliza5/starprod/embedding/P05ic
Xi_102_strange: /eliza5/starprod/embedding/P05ic
Xi_103_strange: /eliza5/starprod/embedding/P05ic
Xi_104_strange: /eliza5/starprod/embedding/P05ic
Xi_105_strange: /eliza5/starprod/embedding/P05ic
Xi_110_strange: /eliza5/starprod/embedding/P05ic
Xi_111_strange: /eliza5/starprod/embedding/P05ic
Xi_112_strange: /eliza5/starprod/embedding/P05ic
Xi_113_strange: /eliza5/starprod/embedding/P05ic
Xi_114_strange: /eliza5/starprod/embedding/P05ic
Xi1530_405_strange: /eliza5/starprod/embedding/P05ic
Xi1530_406_strange: /eliza5/starprod/embedding/P05ic
Xi1530_407_strange: /eliza5/starprod/embedding/P05ic
Xi1530_408_strange: /eliza5/starprod/embedding/P05ic
P04ik:
dbar_101_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_102_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_103_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_104_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_105_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_106_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_107_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_108_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_109_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
dbar_110_200GeV: /auto/pdsfdv37/starprod/embedding/P04ik
Pi0_300_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_301_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_302_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_303_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_304_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_305_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_306_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_307_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_308_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_309_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_310_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_311_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_312_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_313_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_314_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_315_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_316_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_317_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_318_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
Pi0_319_1112996078: /auto/pdsfdv40/starprod/embedding/P04ik
dbar_101_200GeV: /eliza1/starprod/embedding/P04ik
Pi0_282_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_283_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_284_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_285_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_286_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_287_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_288_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_289_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_290_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_291_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_292_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_293_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_294_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_295_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_296_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_297_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_298_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_299_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_311_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_312_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_313_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_314_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_315_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_316_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_317_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_318_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_319_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_321_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_322_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_323_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_324_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_325_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_326_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_327_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_328_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_329_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_331_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_332_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_333_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_334_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_335_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_336_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_337_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_338_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_339_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_340_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_341_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_341_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_342_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_342_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_343_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_343_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_344_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_344_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_345_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_345_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_346_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_346_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_347_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_347_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_348_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_348_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_349_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_349_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_350_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_350_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_351_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_351_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_352_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_352_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_353_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_353_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_354_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_354_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_355_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_355_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_356_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_356_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_357_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_357_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_358_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_358_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_359_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_359_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_360_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_360_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_361_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_361_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_362_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_362_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_363_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_363_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_364_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_364_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_365_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_365_flatpt: /eliza1/starprod/embedding/P04ik P
i0_366_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_366_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_367_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_367_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_368_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_368_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_369_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_369_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_370_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_370_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_371_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_371_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_372_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_372_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_373_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_373_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_374_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_374_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_375_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_375_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_376_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_376_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_377_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_377_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_378_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_378_flatpt: /eliza1/starprod/embedding/P04ik
Pi0_379_dalitz: /eliza1/starprod/embedding/P04ik
Pi0_379_flatpt: /eliza1/starprod/embedding/P04ik
P04ih: P04if:
Pi0_106_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_107_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_108_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_109_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_110_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_111_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_112_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_113_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_114_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_115_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_116_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_120_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_121_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_122_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_123_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_124_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_125_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_127_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_128_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_130_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_141_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_142_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_143_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_144_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_145_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_146_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_147_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_148_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_149_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_150_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_161_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_162_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_163_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_164_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_165_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_166_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_167_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_168_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_169_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_170_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_171_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_172_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_173_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_174_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_175_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_176_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_177_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_178_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_179_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_180_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_182_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_183_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_184_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_185_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_186_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_187_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_188_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_189_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_190_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_191_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_192_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_193_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_194_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_195_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_196_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_197_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_198_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Pi0_199_EMC: /auto/pdsfdv37/starprod/embedding/P04if
Sigma1385barminus_501_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barminus_502_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barminus_503_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barminus_504_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barminus_505_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barplus_502_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barplus_503_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barplus_504_heavy: /eliza5/starprod/embedding/P04if
Sigma1385barplus_505_heavy: /eliza5/starprod/embedding/P04if
Sigma1385minus_501_heavy: /eliza5/starprod/embedding/P04if
Sigma1385minus_502_heavy: /eliza5/starprod/embedding/P04if
Sigma1385minus_503_heavy: /eliza5/starprod/embedding/P04if
Sigma1385minus_504_heavy: /eliza5/starprod/embedding/P04if
Sigma1385minus_505_heavy: /eliza5/starprod/embedding/P04if
Sigma1385plus_501_heavy: /eliza5/starprod/embedding/P04if
Sigma1385plus_502_heavy: /eliza5/starprod/embedding/P04if
Sigma1385plus_503_heavy: /eliza5/starprod/embedding/P04if
Sigma1385plus_504_heavy: /eliza5/starprod/embedding/P04if
Sigma1385plus_505_heavy: /eliza5/starprod/embedding/P04if
P04ie:
Lambda_301Svt_62GeV: /auto/pdsfdv45/starprod/embedding/P04ie
Xi_201_62GeV: /auto/pdsfdv65/starprod/embedding/P04ie
Xi_202_62GeV: /auto/pdsfdv65/starprod/embedding/P04ie
Xi_203_62GeV: /auto/pdsfdv65/starprod/embedding/P04ie
K0Short_201_62GeV: /eliza1/starprod/embedding/P04ie
K0Short_202_62GeV: /eliza1/starprod/embedding/P04ie
K0Short_203_62GeV: /eliza1/starprod/embedding/P04ie
K0Short_211_62GeV: /eliza1/starprod/embedding/P04ie
Lambda_211_62GeV: /eliza1/starprod/embedding/P04ie
P03ih:
Pi0_502_dAu: /eliza5/starprod/embedding/P03ih
P03if:
TEST: /eliza1/starprod/embedding/P03if
test_sigma: /eliza1/starprod/embedding/P03if
P03ie: P03id: P02ge: P02gd: Xi_noAcc_101_Central: /eliza1/starprod/embedding/P02gd K0Short_P02gd_101_Minbias: /eliza5/starprod/embedding/P02gd K0Short_P02gd_102_Minbias: /eliza5/starprod/embedding/P02gd K0Short_P02gd_103_Minbias: /eliza5/starprod/embedding/P02gd
LPP group (Dubna) activities
This page is intended to provide embedding progress for soft-photon study conducted by PPL (Dubna, JINR) group in Russia.- Requested embedding is based on request ID # 1103209240 and modified to:
- provide larger statistics for reco efficiency calculation
- extend acceptance parameters
- Related embedding jobs currently in production:
- ~420 kEvents done and loaded on the disk
- This is the embedding request and progress detals
- AuAu200, P05ic:
- Related embedding jobs to be planned:
- Jobs have not been started yet
- This is the embedding request and progress detals
- dAu200, P04if
AuAu200 photon embedding details
- Request details:
- Dataset and production: AuAu200, P05ic
- Embedded particles and multiplicity: 1000 Photons per event, total 500 kEvents
- zVertex = +-30
- Pt: 0.020 - 0.160 GeV
- eta: +-1.2
- full geant reconstruction
- Embedding Files on Disk at PDSF:
- Photon_201...215_spectra: /eliza5/starprod/embedding/P05ic/
- Photon_301...315_spectra: /eliza5/starprod/embedding/P05ic/
- Photon_401...415_spectra: /eliza7/starprod/embedding/P05ic/
- Photon_501...504_spectra: /eliza5/starprod/embedding/P05ic/
- Photon_505...515_spectra: /eliza7/starprod/embedding/P05ic/
- Photon_601...615_spectra: /eliza7/starprod/embedding/P05ic/
- Photon_701...715_spectra: /eliza12/starprod/embedding/P05ic/
- Photon_802...811,817,818_spectra: /eliza12/starprod/embedding/P05ic/
- Running Jobs status:
jobs QA and checking if this statistic is enough
dAu photon embedding details
- Request details:
- Dataset and production: dAu200, P04if
- Embedded particles and multiplicity: 1000 Photons per event, total 500 kEvents
- zVertex = +-50
- Pt: 0.020 - 0.160 GeV
- eta: +-1.2
- full geant reconstruction
- Running Jobs status:
have not been started yet
QA Status (OBSOLETE)
The full list of STAR embedding requests (Since August, 2010):http://drupal.star.bnl.gov/STAR/starsimrequest
The QA status of each request can be viewed in its history.
The information below are only valid for OLD embedding requests.
This document is intended to provide all the information available for each production, starting for the last or current production.
P06ib
K star
Files: pdsf>/eliza1/starprod/embedding/KstarM* ( July 31 2007)Plots for Dedx, Mips, eta, phi distributions and vertex position on K star embedding are attached. Just around 20% of K star Minus files could be scanned due to space issues (in my home dir) at pdsf. Reconstructed tracks were done just on Daugther Pions. There is little difference(0.036) in the mips position possible due to lack of statistics.
Plots - Dca Distributions
The Following are the results for dca distributions for different pT bins in Pi Minus. The results of dca agrees reasonable for the cut-off of 2 cm and above. These plots were done using the macro plot_dca.C
Event Selection Criteria
* Production: P06ib
* Vertex restriction: |x|<3.5cm, |y|<3.5cm, |z|<25.0cm
* N Fit points >= 25
* DCA < 3 cm
- PiMinus
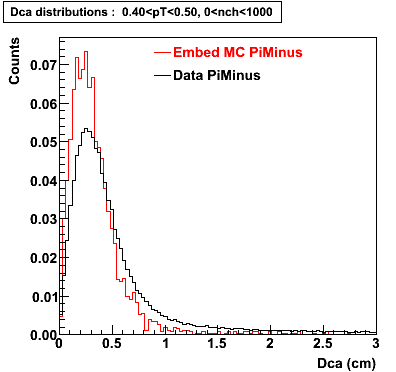
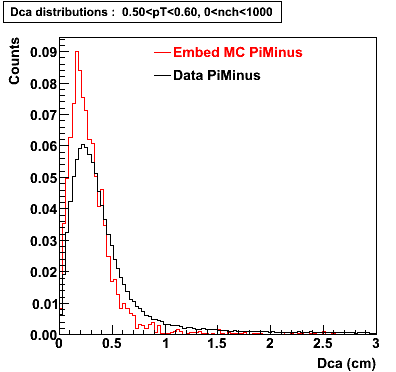
0.20<pT<0.30 0.30<pT<0.40 0.40<pT<0.50 0.50<pT<0.60 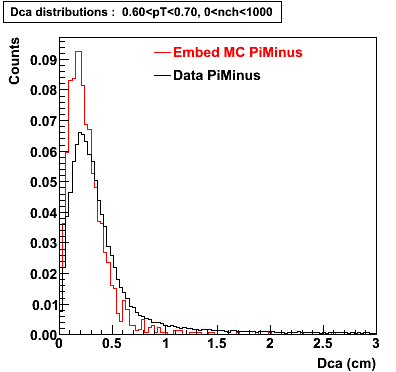
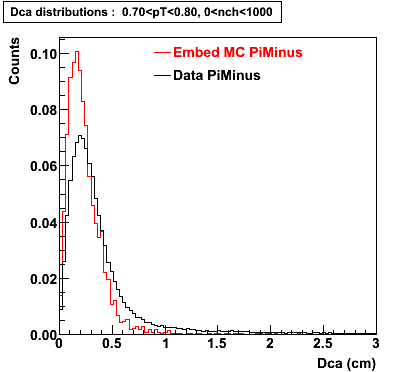
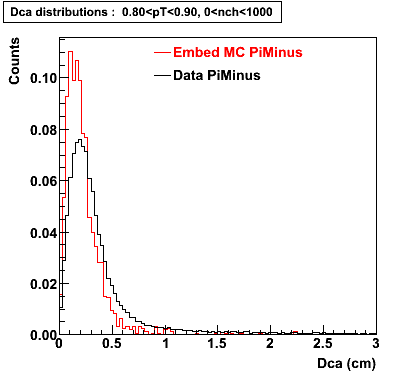
0.60<pT<0.70 0.70<pT<0.80 0.80<pT<0.90 0.90<pT<1.0
PiPlus
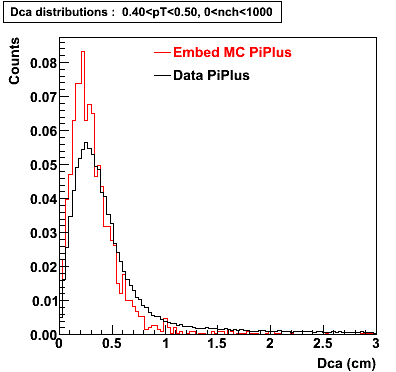
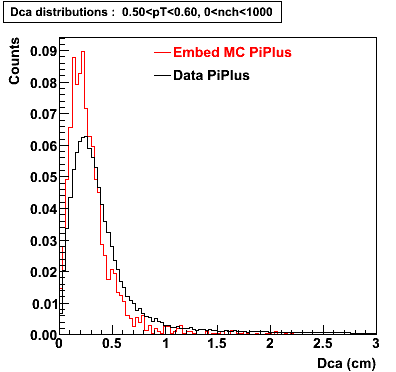
0.20<pT<0.30 0.30<pT<0.40 0.40<pT<0.50 0.50<pT<0.60 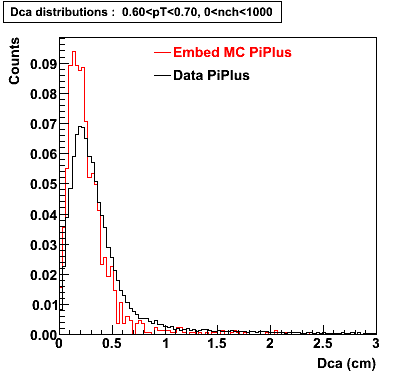
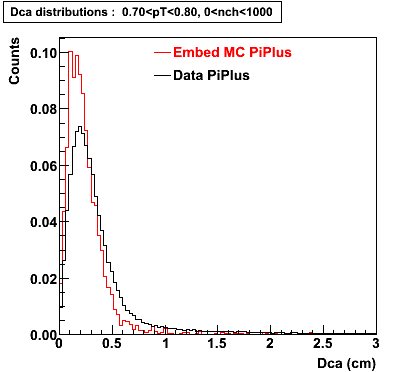
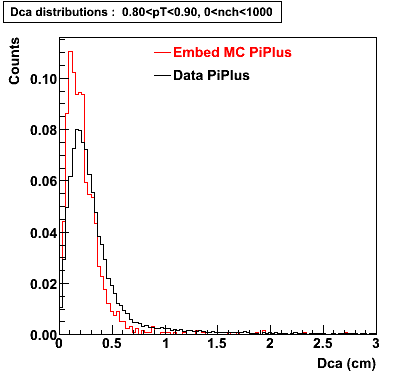
0.60<pT<0.70 0.70<pT<0.80 0.80<pT<0.90 0.90<pT<1.0
K Minus
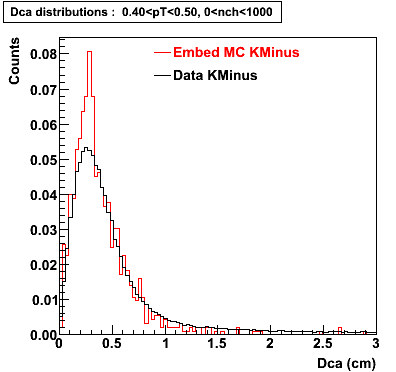
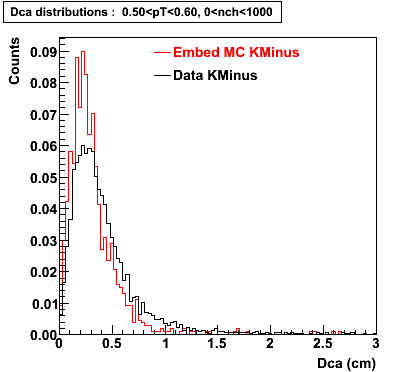
0.20<pT<0.30 0.30<pT<0.40 0.40<pT<0.50 0.50<pT<0.60 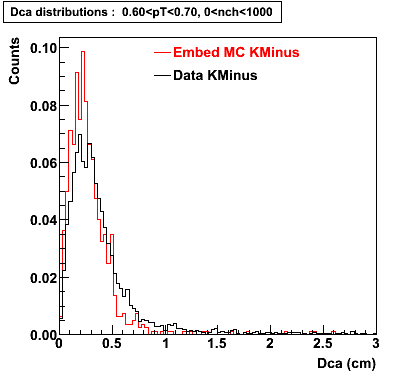
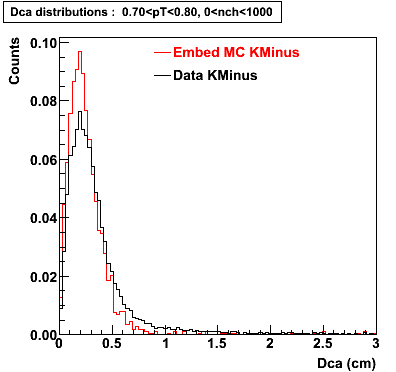
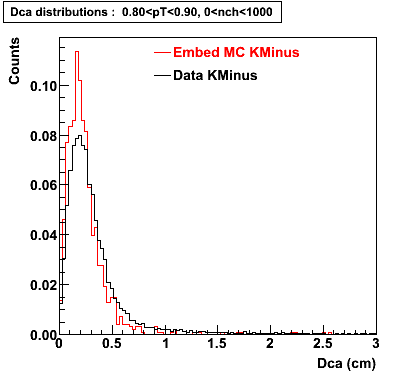
0.60<pT<0.70 0.70<pT<0.80 0.80<pT<0.90 0.90<pT<1.0
K Plus
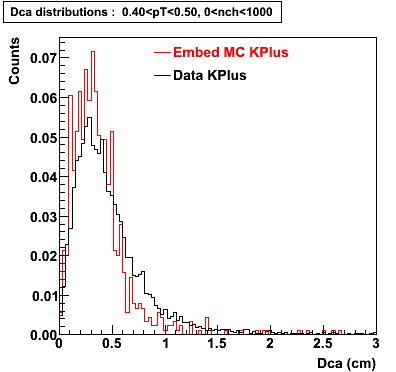
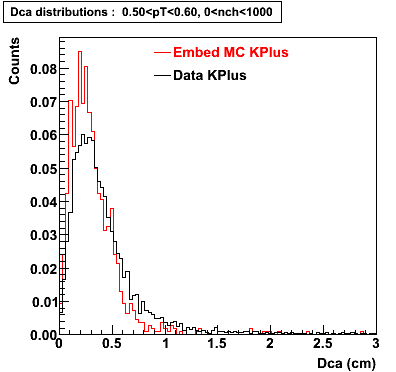
0.20<pT<0.30 0.30<pT<0.40 0.40<pT<0.50 0.50<pT<0.60 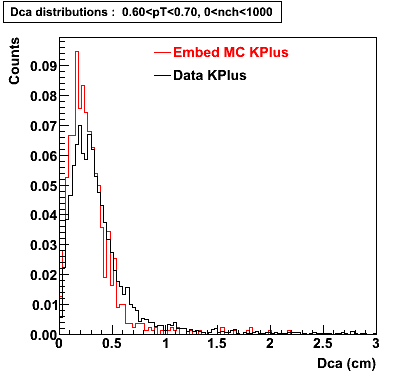
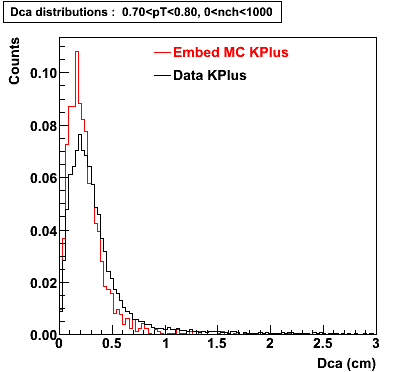
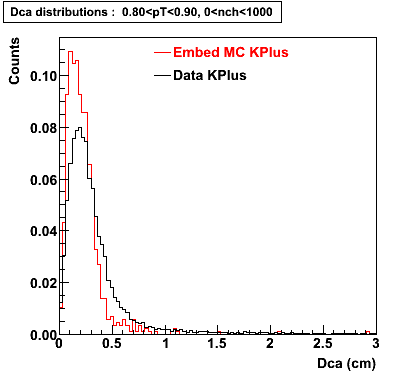
0.60<pT<0.70 0.70<pT<0.80 0.80<pT<0.90 0.90<pT<1.0
Proton
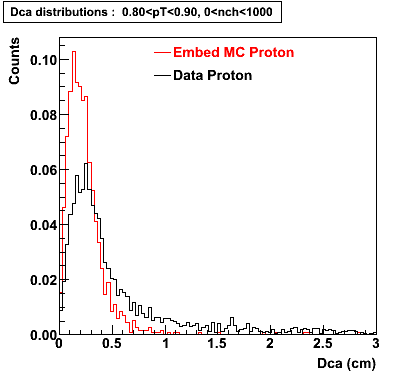
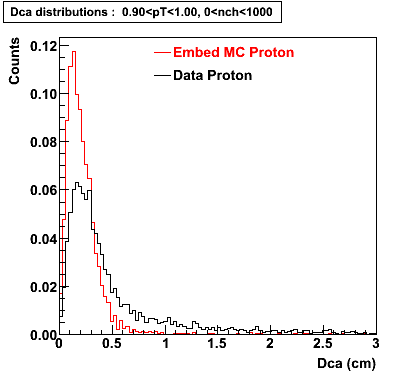
0.40<pT<0.50 0.50<pT<0.60 0.80<pT<0.90 0.90<pT<1.00
P bar
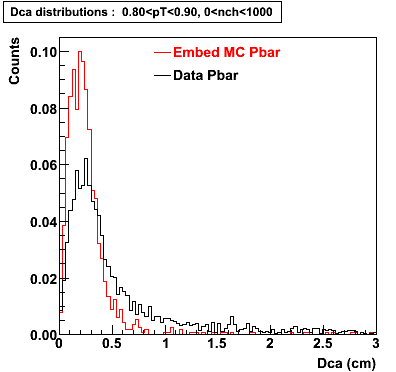
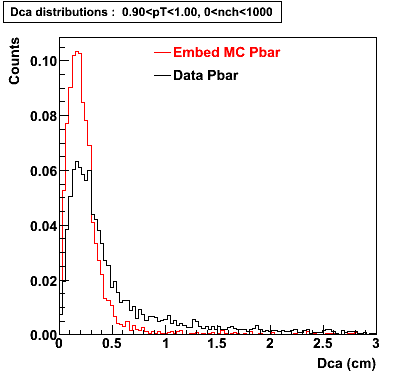
0.40<pT<0.50 0.50<pT<0.60 0.80<pT<0.90 0.90 < pT < 1.00
Plots - Number of Fitted Points
The Following are the results for dca distributions for different pT bins in Pi Minus. The results of dca agrees reasonable for the cut-off of 2 cm and above. These plots were done using the macro plot_dca.CEvent Selection Criteria
* Production: P06ib
* Vertex restriction: |x|<3.5cm, |y|<3.5cm, |z|<25.0cm
* N Fit points >= 25
* DCA < 3 cm
- Pi Minus
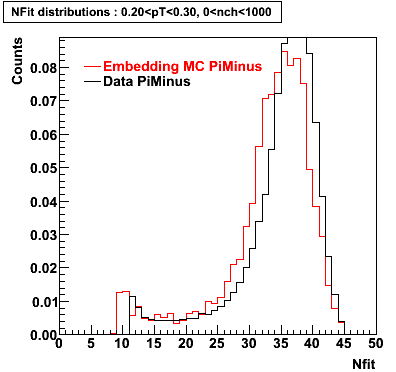 | 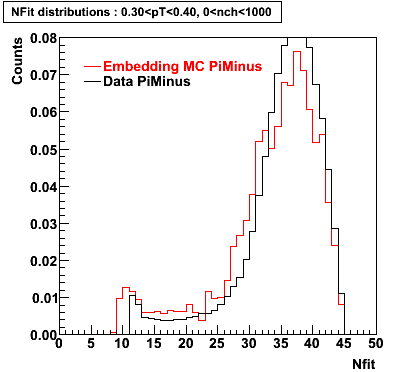 | 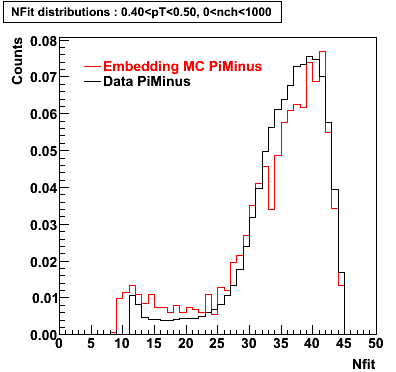 | 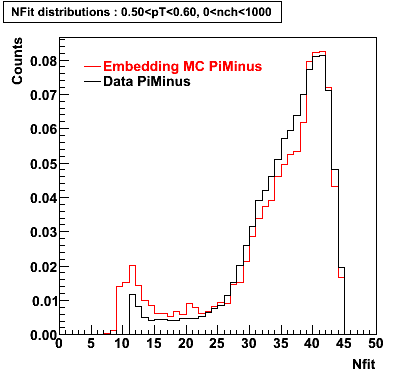 |
| 0.20 < pT < 0.30 | 0.30 < pT < 0.40 | 0.40 < pT < 0.50 | 0.50 < pT < 0.60 |
- Pi Plus
Pi Minus
|
||
|
||
|
||
|
||
|
Pi Plus
|
||
|
||
|
||
|
||
|
K Minus
|
||
|
||
|
||
|
||
|
K Plus
|
||
|
||
|
||
|
||
|
Proton
|
||
|
||
|
||
|
||
|
P bar
|
||
|
||
|
||
|
||
|
QA Plots Phi(Feb 2009)
QA P06ib (Phi->K +K)
This is the QA for P06ib (Phi- > KK). reconstruction on Global Tracks (Kaons)
1. dEdx
Reconstruction on Kaon Daugthers. Plot shows MOntecarlo tracks and Ghost Tracsks.
/P06ib_dedx_Phi(Ghost).gif)
2. DCA Distributions
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots. (MonteCarlo and MuDst) (MuDst taken from pdsf > /eliza12/starprod/reco/cuProductionMinBias/ReversedFullField/P06ib/2005/022/st_physics_adc_6022048_raw*.MuDst.root)
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
/dca_eta_pt_phi.gif)
3. NFit Distributions
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions. (MonteCarlo and MuDst)
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
/nfit_eta_pt_phi.gif)
4. Delta Vertex
The following are the Delta Vertex ( Vertex Reconstructed - Vertex Embedded) plot for the 3 diiferent coordinates (x, y and z) (Cuts of vz =30 cm , NFitCut= 25 are applied)
/Delta_vertex_Phi.gif)
5. Z Vertex and X vs Y vertex
/vertex_Phi.gif)
6. Global Variables : Phi and Rapidity
/y_f_phi(1).gif)
/phi_f_phi.gif)
7. Pt
Embedded Phi meson with flat pt (black)and Reconstructed Kaon Daugther (red).
/pt_f_phi.gif)
8. Randomness Plots
The following plots, are to check the randomness of the input Monte Carlo (MC) tracks.
/mc_pt_y_Phi_K.gif)
/mc_y_phi_phi_K.gif)
QA plots Rho (February 16 2009)
QA P06ib (Rho->pi+pi)
This is the QA for P06ib (Rho- > pi+pi). reconstruction on Global Tracks (pions)
1. dEdx
Reconstruction on Pion Daugthers.

2. DCA Distributions
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
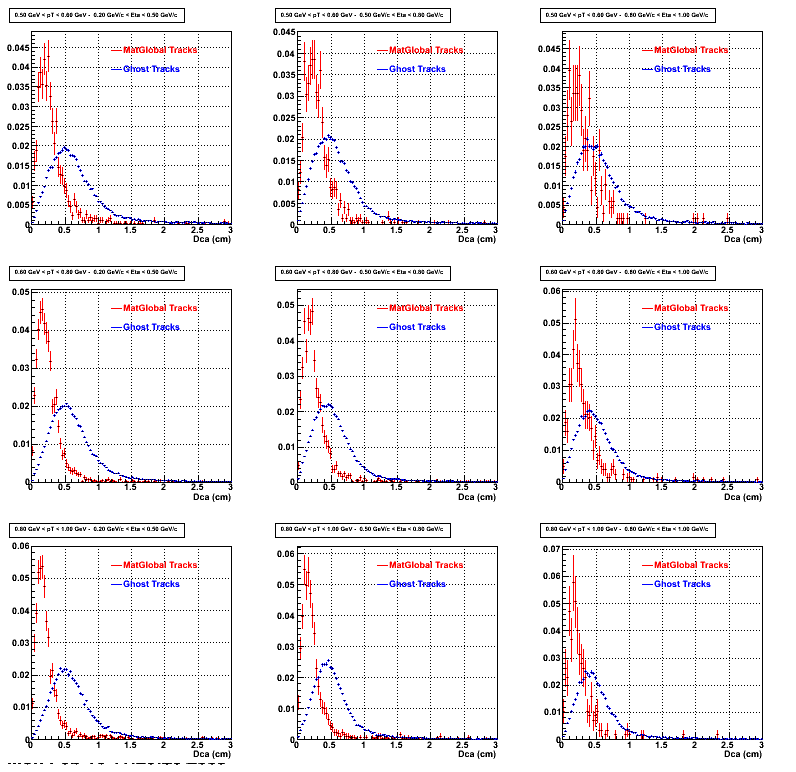
2b. Compared with MuDst
.gif)
3. NFit Distributions
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
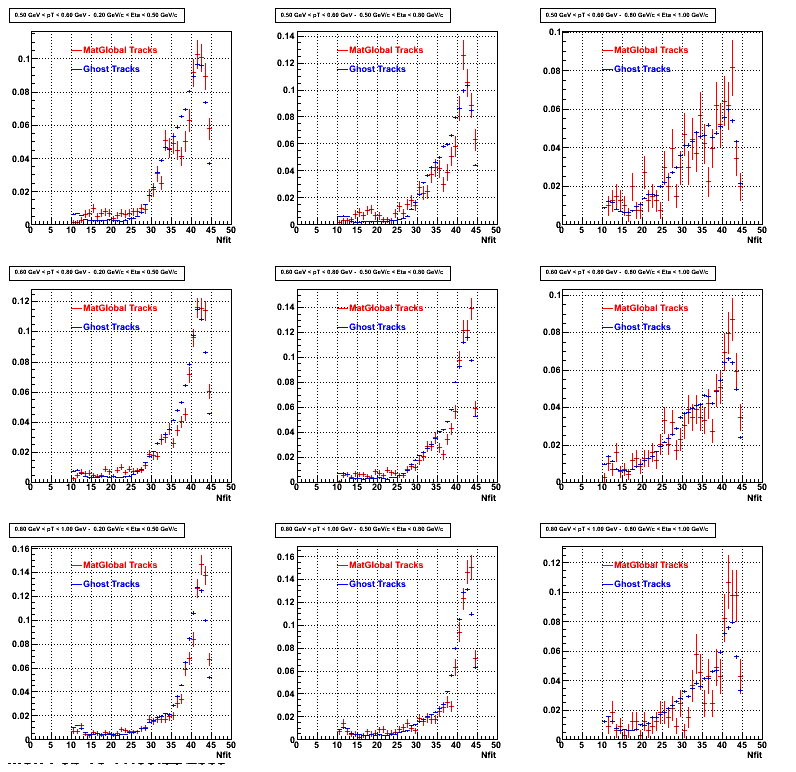
3b. Reconstructed compared with MuDsts
.gif)
4. Delta Vertex
The following are the Delta Vertex ( Vertex Reconstructed - Vertex Embedded) plot for the 3 diiferent coordinates (x, y and z) (Cuts of vz =30 cm , NFitCut= 25 are applied)
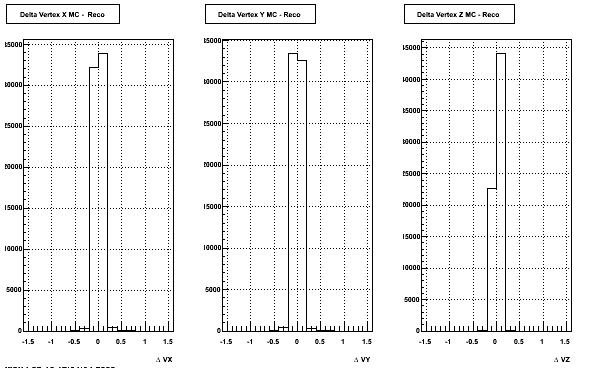
5. Z Vertex and X vs Y vertex
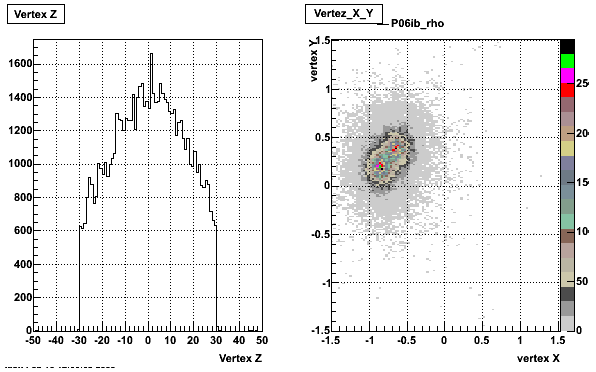
6. Global Variables : Phi and Rapidity
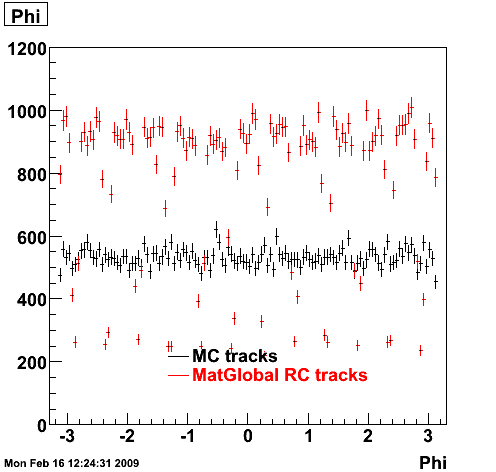
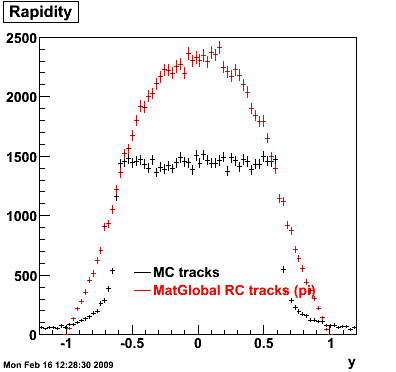
7. Pt
Embedded Rho meson with flat pt (black)and Recosntructed Pion (red).
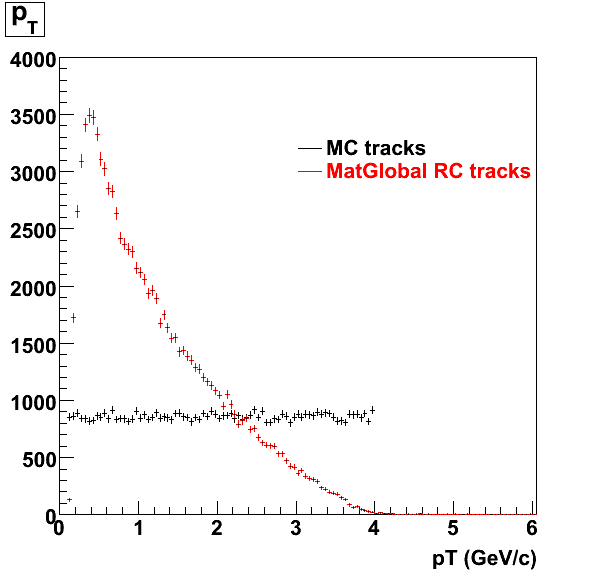
8. Randomness Plots
The following plots, are to check the randomness of the input Monte Carlo (MC) tracks.
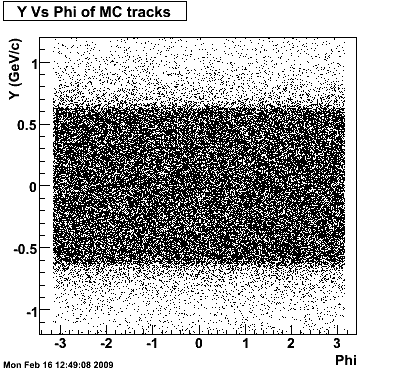
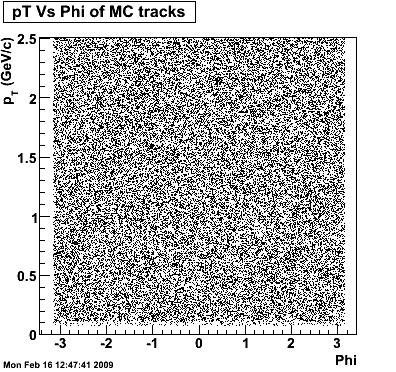
.gif)
QA plots Rho (October 21 2008)
Some QA plots for Rho:
MiniDst files are at PDSF under the path /eliza13/starprod/embedding/p06ib/MiniDst/rho_101/*.minimc.root
MuDst files are at PDSF under /eliza13/starprod/embedding/P06ib/MuDst/10*/*.MuDst.root Reconstruction had been done on PionPlus.
DCA and Nfit Distributions had been scaled by the Integral in different pt ranges
QAPlots_D0
Some QA Plots for D0 located under the path :
/eliza12/starprod/embedding/P06ib/D0_001_1216876386/*
/eliza12/starprod/embedding/P06ib/D0_002_1216876386/ -> Directory empty
Global pairs are used as reconstructed tracks. SOme quality tractst plotiing level were :
Vz Cut 30 cm ;
NfitCut : 25,
Ncommonhits : 10 ;
maxDca :1 cm ; Assuming D0- >pi + pi
P07ib
Test done on P07ib - Doing claibration on dE/dx |
 |
Fraction bla bla
 |
P05id
P05id Cu+Cu Embedding
Hit level check-up : At the Hit level the results look in good agreement.
- Missing/Dead Areas (PiMinus): The next graphs show dead sectors for embeded data and real data as well.
- Missing/Dead Areas (PiPlus)
- Track Residuals PiMinus
- Track Residuals PiPlus
- dEdx Comparisons
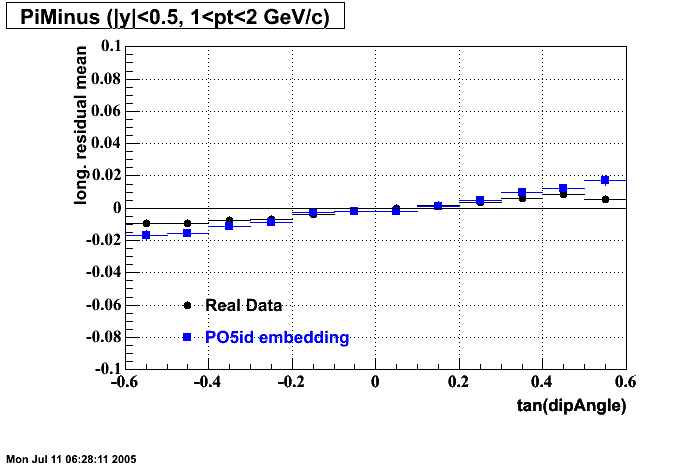 | |
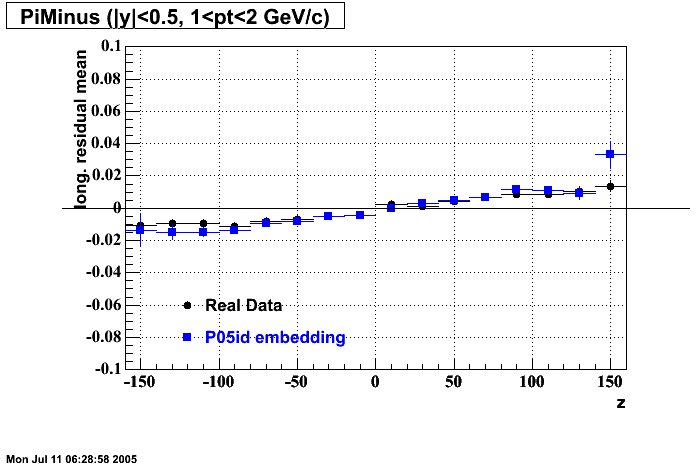 | |
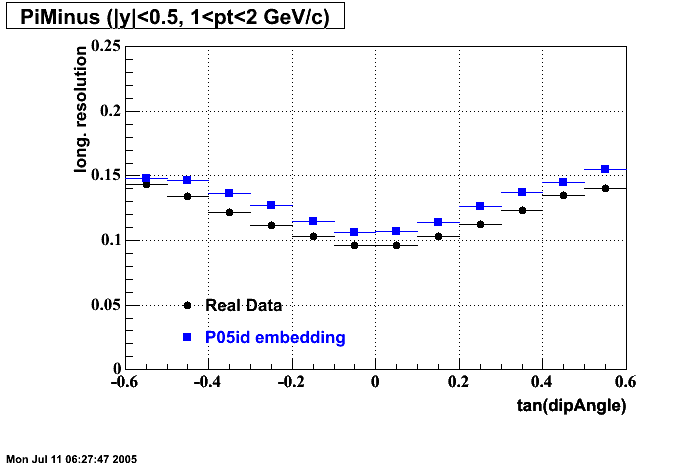 | |
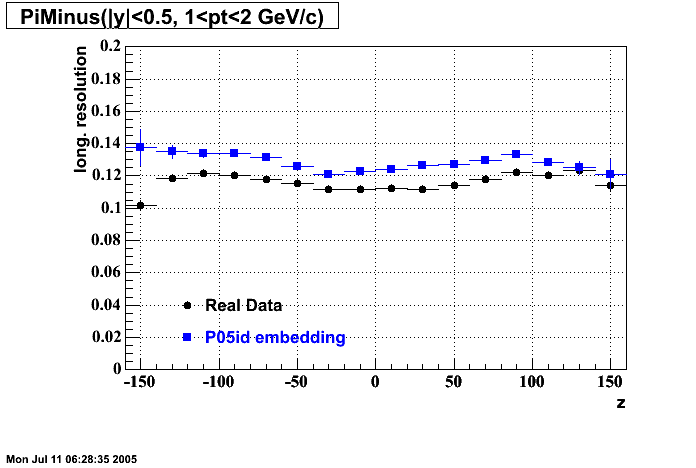 | |
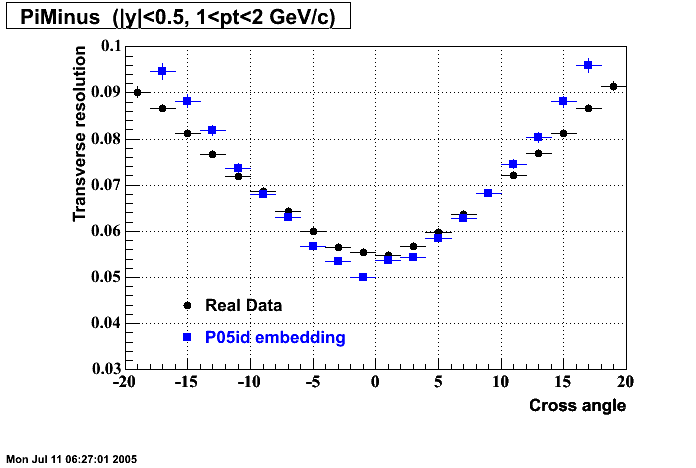 | |
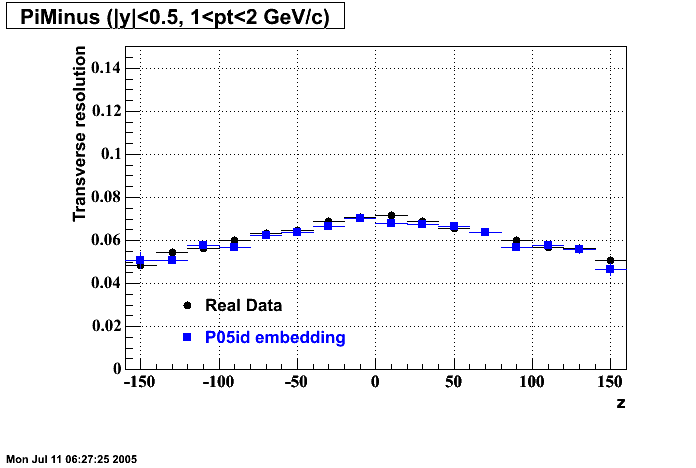 |
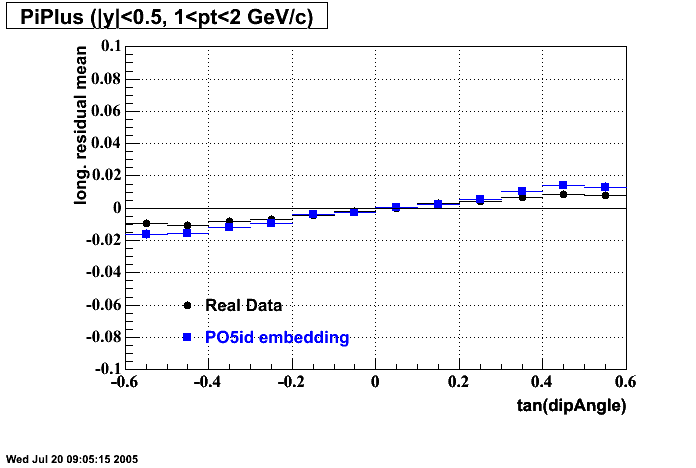 | |
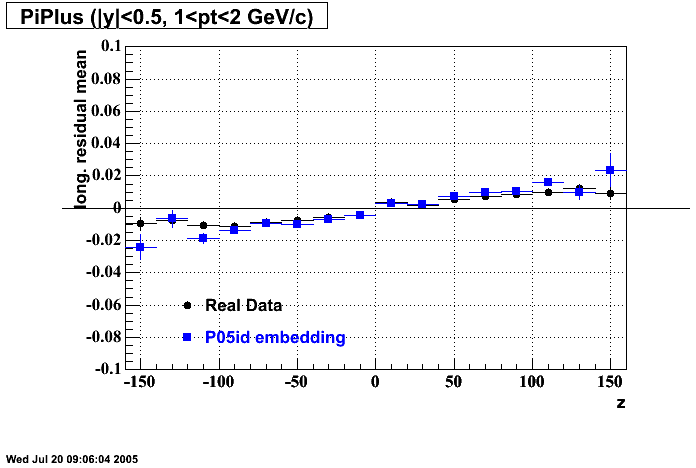 | |
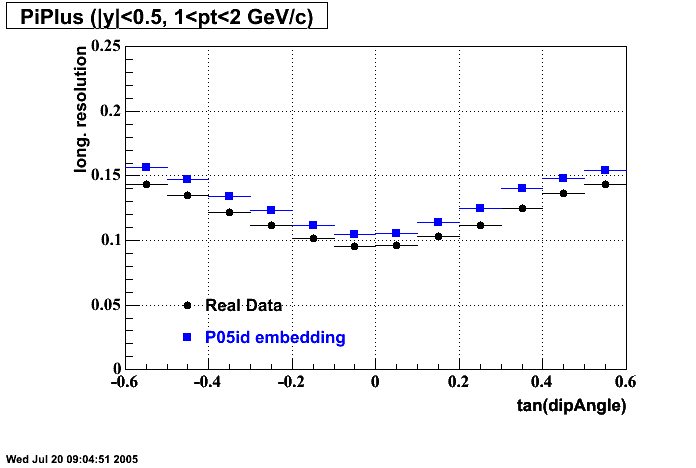 | |
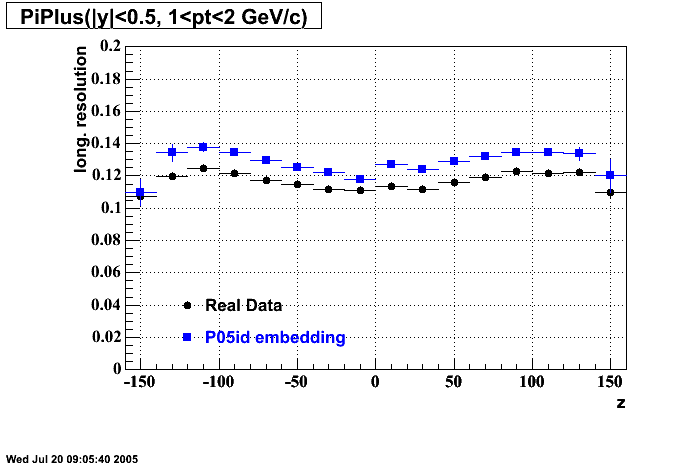 | |
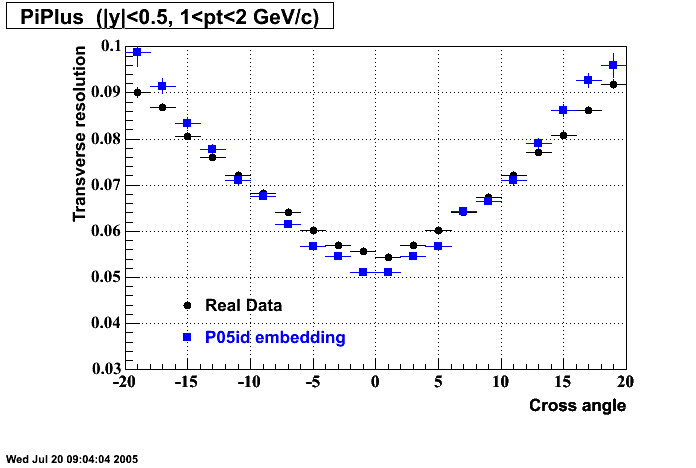 | |
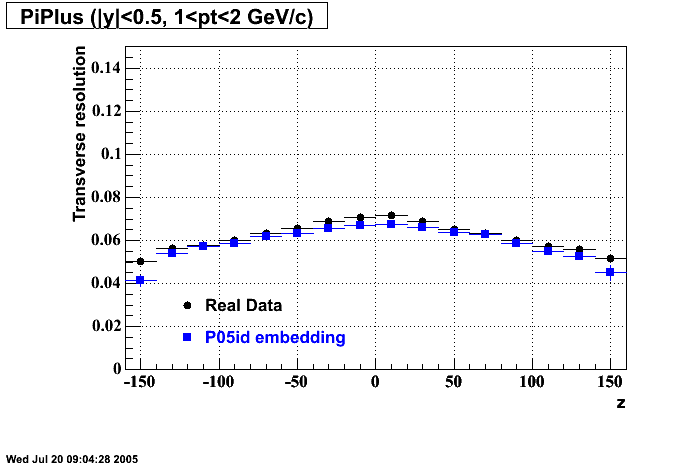 |
The following are results of dEdx calibration from embedding for Pi Minus. Graphs 1 and 2 are Dedx vs Momentum grapsh where Green dots are the MonteCarlo Tracks reconstructed from Embedding and Black dots are Data. In Graph Number 2, dots are Data and solid lines show Bichsel parametrisation with a factor of 1/2 offset. In the Last Graph a projection on dEdx axis is done and a MIP of 1.26 and 1.18 are shown.
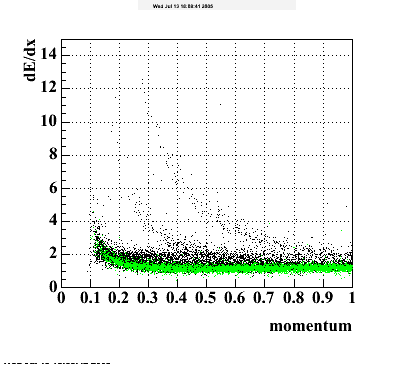 | |
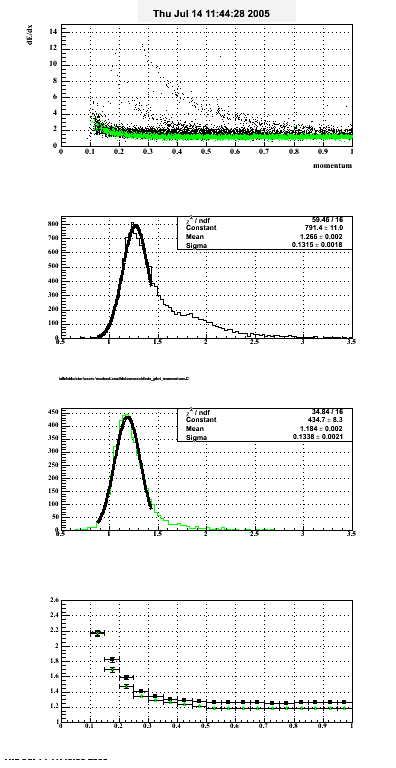 |
P08ic -Jpsi(Test)
QA P08ic J/Psi -> ee+
This is the QA for P08id (jPsi - > ee). Reconstructin on Global Tracks and just Electrons (Positrons).
1. dEdx
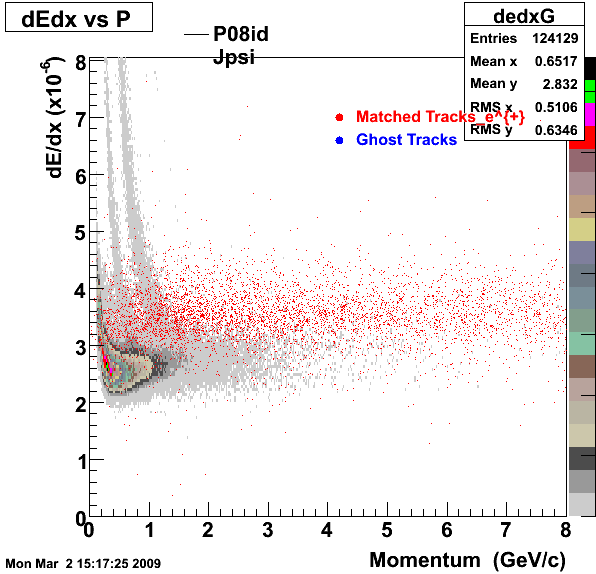
2. DCA Distributions
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();

3. NFit Distributions
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
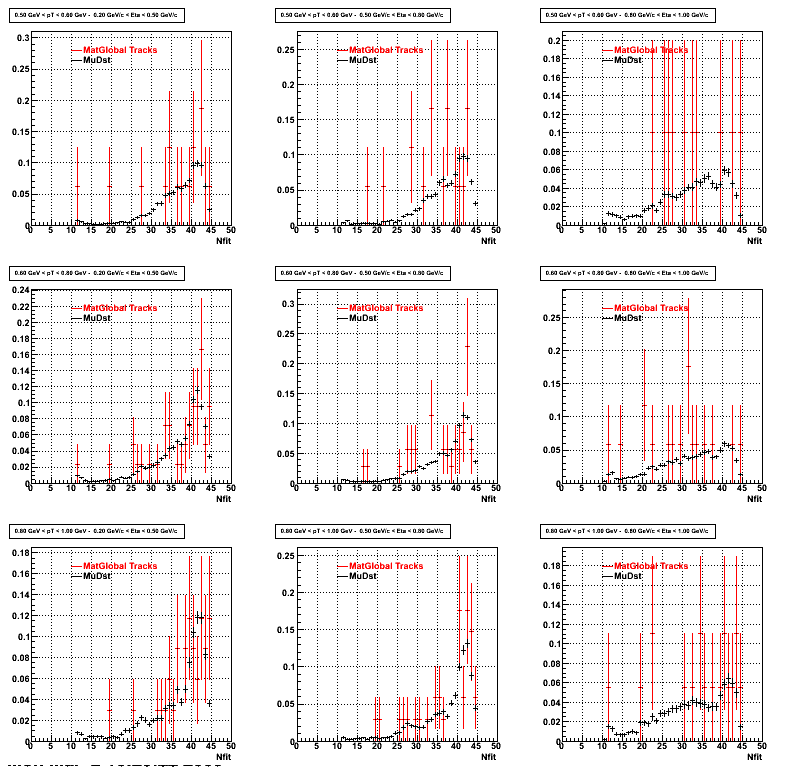
4. Delta Vertex
The following are the Delta Vertex ( Vertex Reconstructed - Vertex Embedded) plot for the 3 diiferent coordinates (x, y and z)
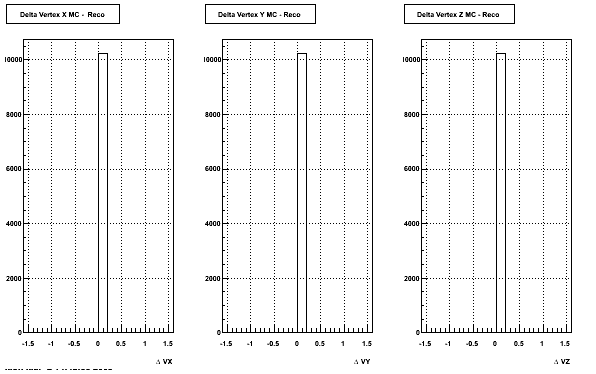
5. Z Vertex and X vs Y vertex
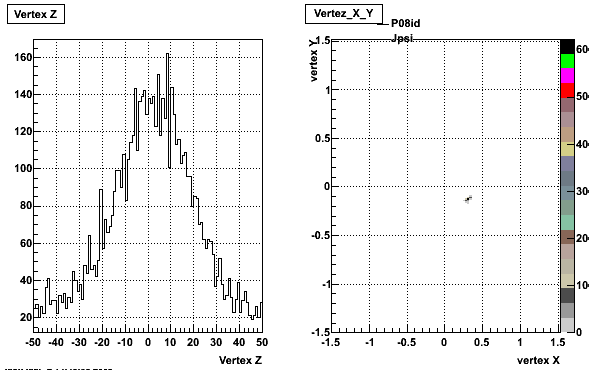
6. Global Variables : Phi and Rapidity
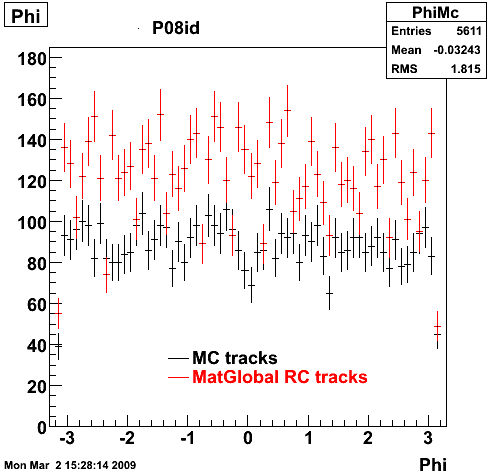
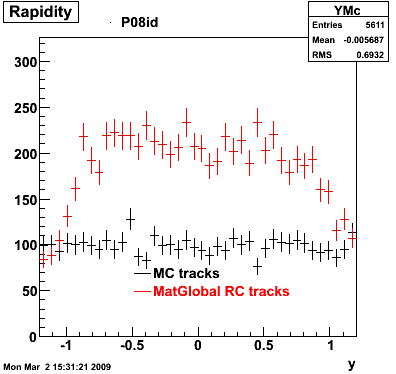
7. Pt
Embedded J/Psi with flat pt (black)and Recosntructed Electrons (red).
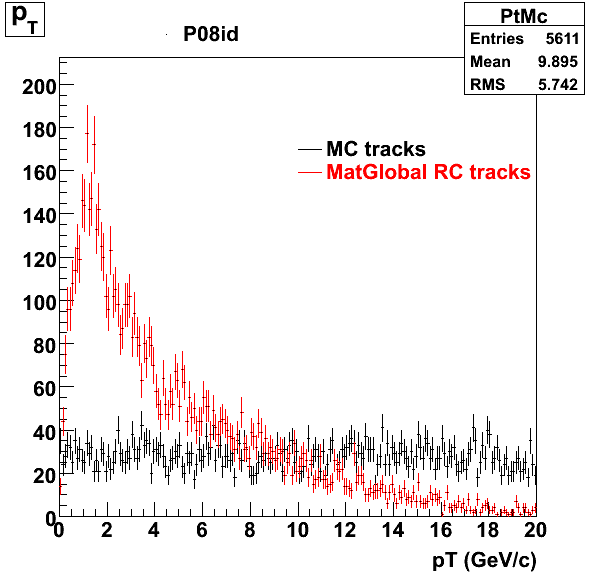
8. Randomness Plots
The following plots, are to check the randomness of the input Monte Carlo (MC) tracks.

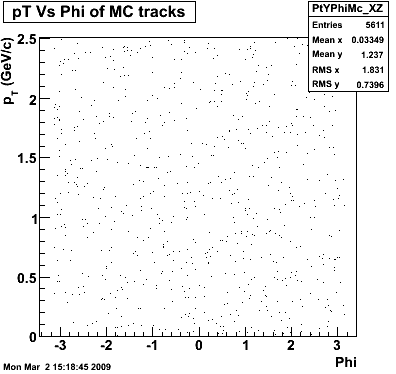
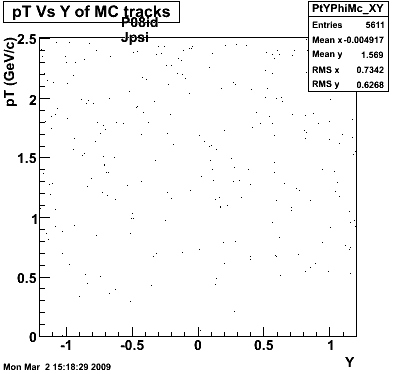
P08id (AXi -> RecoPiPlus)
AXi-> Lamba + Pion + ->P + Pion - + Pion +
(03 08 2009)
1. Dedx
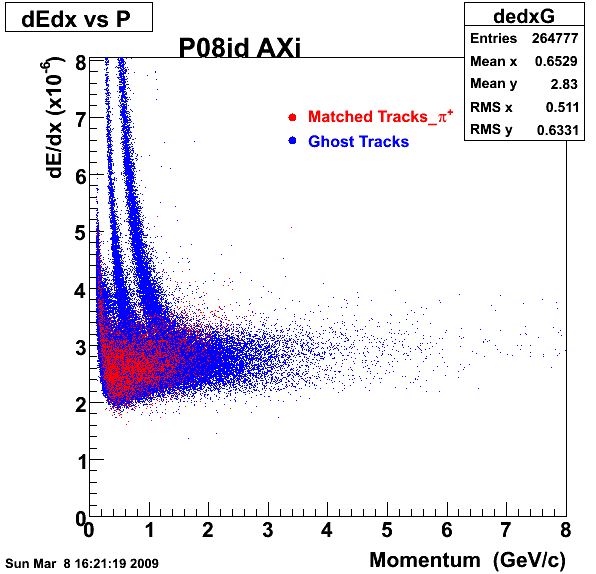
2.Dca
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
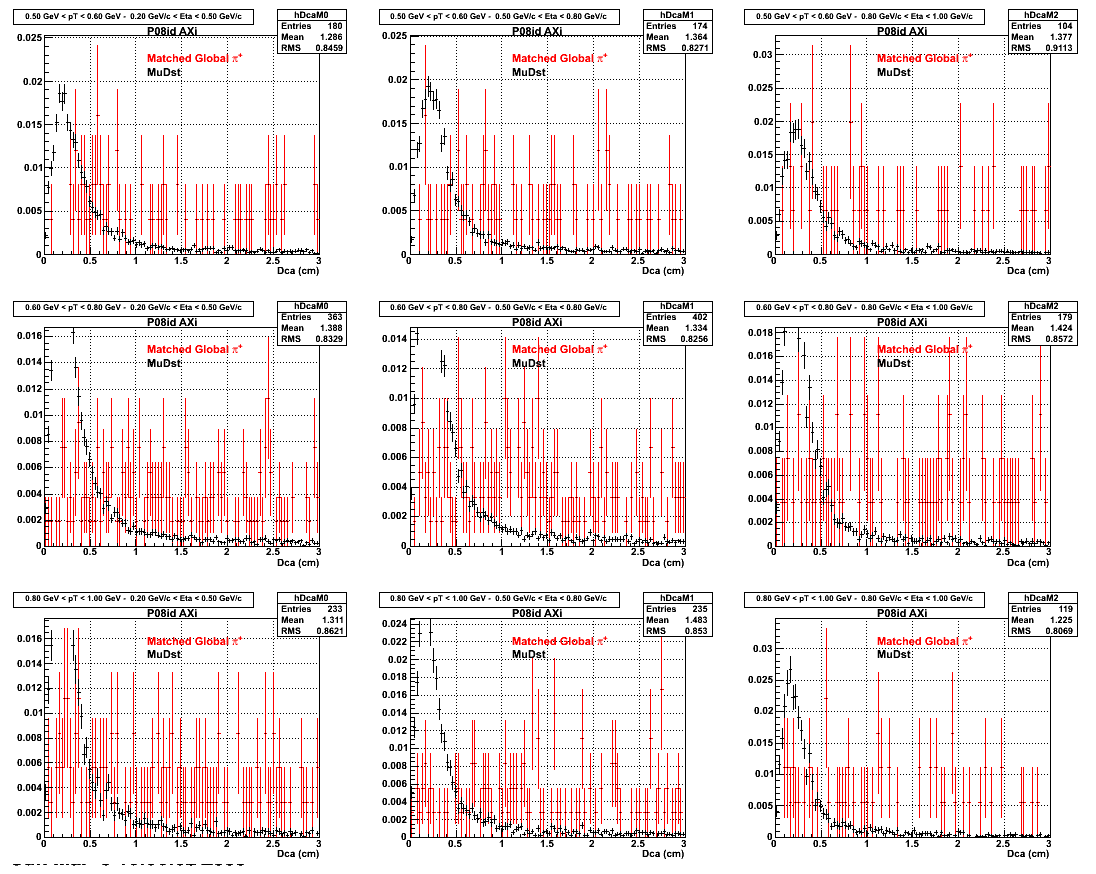
3. Nfit
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
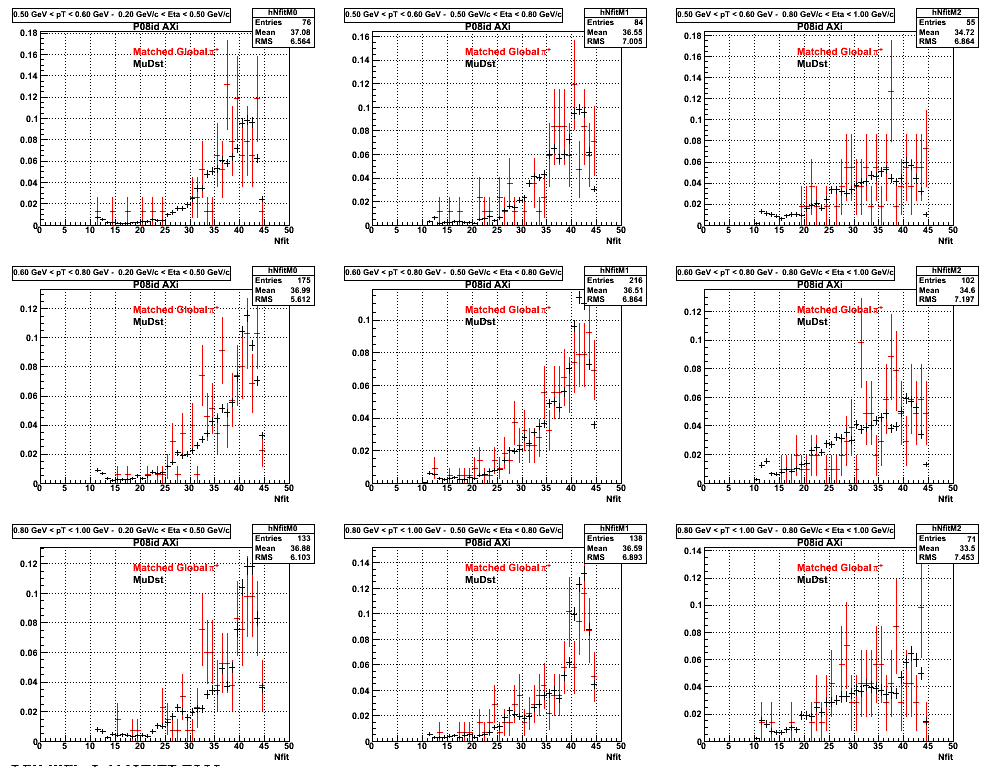
4. Delta Vertex
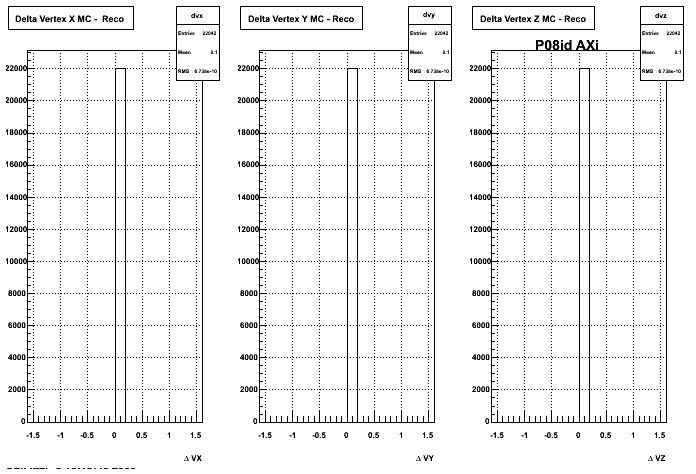
5. Z Vertex and X vs Y vertex

6. Global Variables : Phi and Rapidity
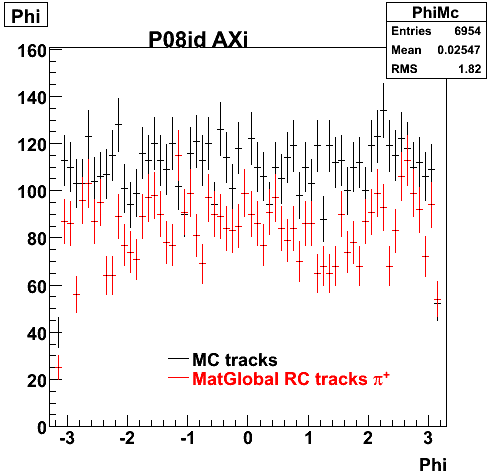
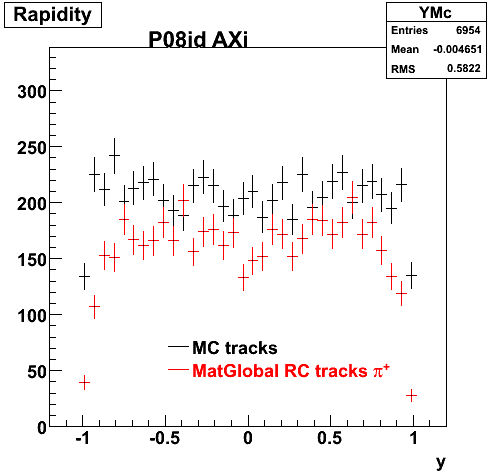
7. pt
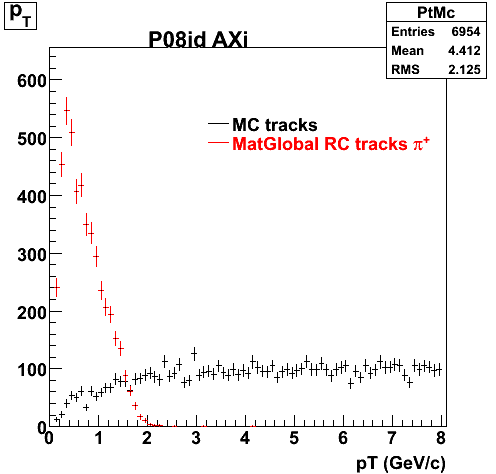
8. Randomness

P08id (Lambda->PK)
QA of Lambda Embedding with run 8 d+Au on PDSF (sample 15x)
Let's first check some event wise information. They look fine.

Then we check the randomness of the input Monte Carlo (MC) Lambda tracks. The 'phasespace' command in GSTAR is used for sampling the MC tracks. The input is supposed to be flat for pT within [0,8], Y [-1.5,1.5] and Phi [-Pi,Pi]. The 3 plots below show the randomness is OK for this sample. Please notice that Y is rapidity, not pseudo-rapidity.



Then we compare the dedx of reconstructed MC (matched) global tracks (i.e. the daughters of MC Lambda) to those of real (or ghost) tracks, to fix the scale factor. (scale factor = 1.38 ?)

Now we compare the nFitHits distribution of matched global tracks (i.e. the daughters of MC Lambda) and real tracks. The cuts are |eta|<1, nFitHits>25. For matched tracks, nCommonHits>10 cut is applied. From the left plot, we can see, the agreement of nHitFits is good for all pT ranges.

We check the pT, rapdity and Phi distributions of reconstruced (RC) Lambda and input (MC) Lambda. The cut for Lambda reconstruction is very loose. They look normal.



Here, we compare some cut variables from the reconstructed (RC) Lambda to those from real Lambda. Again, as you can see in these plots, the cuts are very loose for Lambda (contribution of background is estimated with rotation method, and has been subtracted). These plots are made for 8 pT bins (with rapidity cut |y|<0.75). The most obvious difference is in DCA of V0 to PV, especially for high pT bin.







P08id (Omega -> RecoPiMinus)
Omega-> Lamba + K - -> P + Pion - + K -
(03 08 2009)
1. Dedx
Reconstruction on pion Minus and Proton Daugthers. 2 different plots are shown just for the sake of completenees.... Reconsructing on Kaon had very few statistics
| Reco PionMinus | Reco Proton |
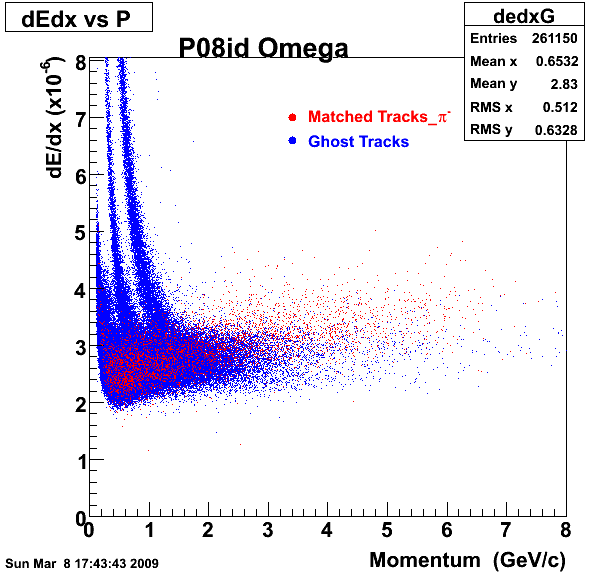 |  |
2.Dca
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
*.Reconstructing on Pion
*. Reconstructing on Proton
3. Nfit
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
*. Reconstructing Pion
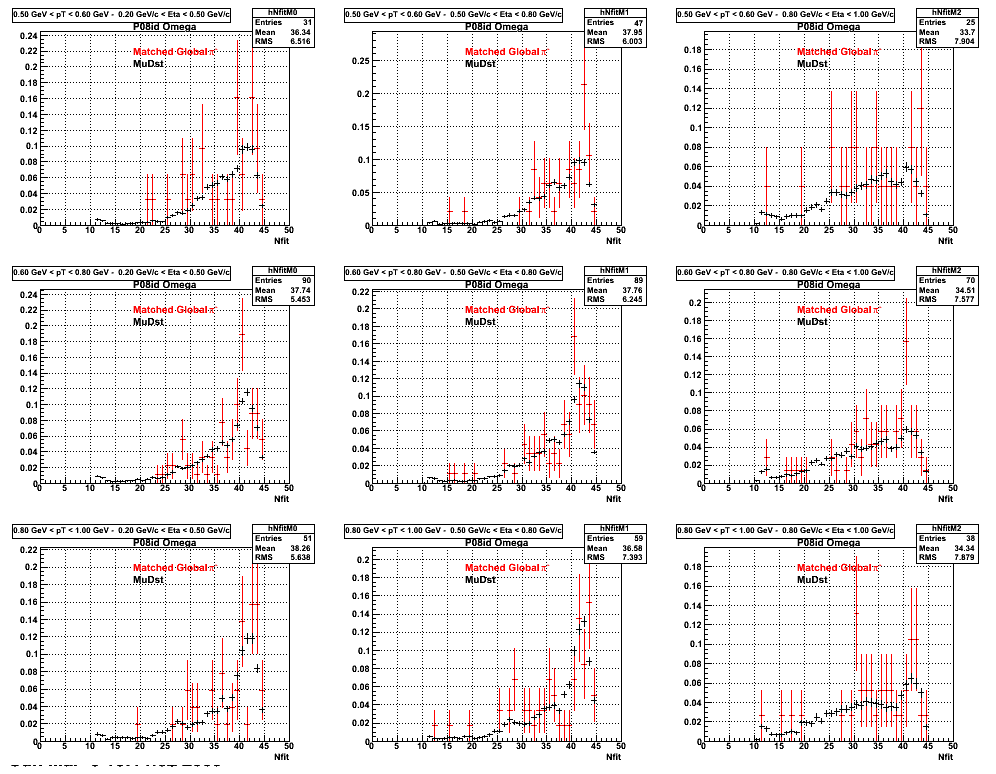
*. Reconstructing Proton
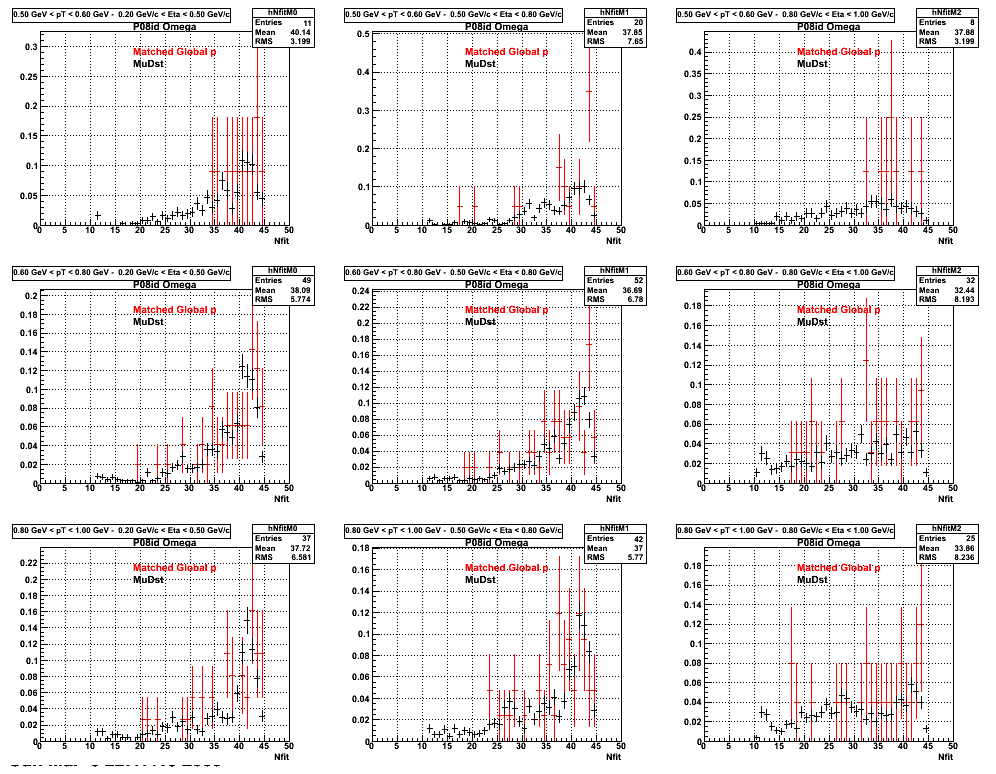
4. Delta Vertex
When reconstructed in Pion Minus and Proton it turns out to have the same ditreibutions so I just posted one of them
5. Z Vertex and X vs Y vertex
6. Global Variables : Phi and Rapidity
| Reco Pion | Reco Kaon |
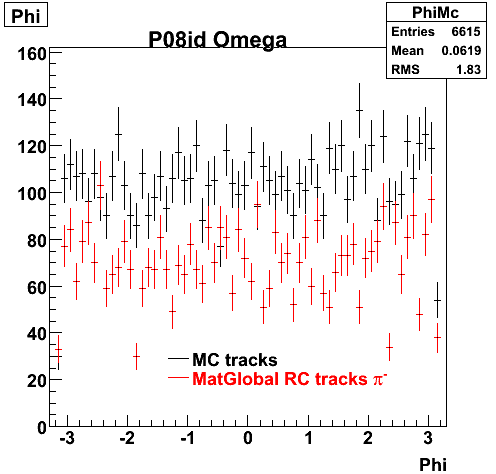 | 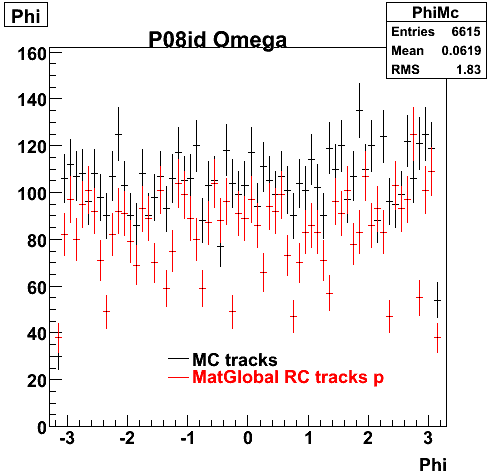 |
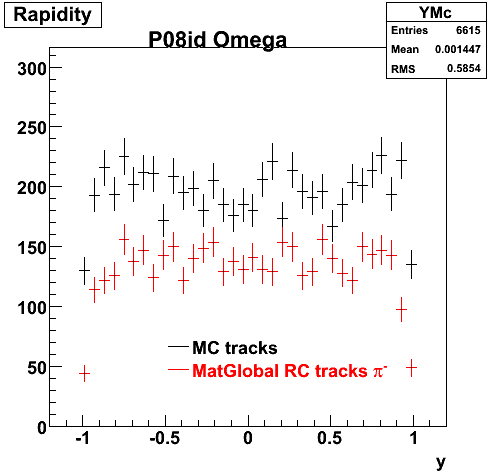 | 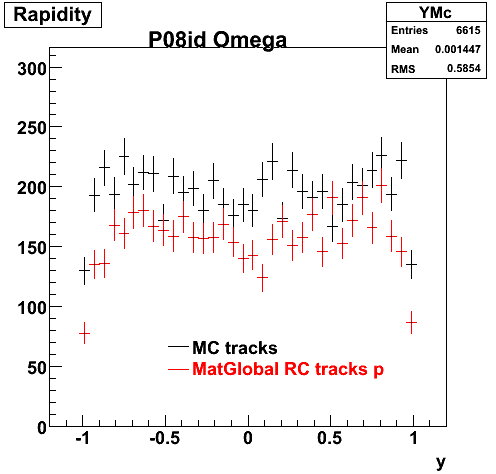 |
7. pt
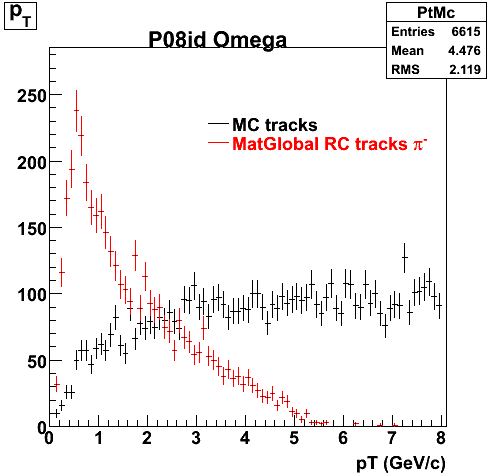
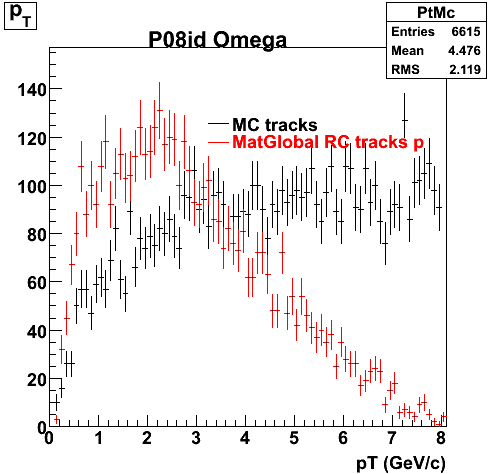
8. Randomness

P08id (phi ->KK) (March 05 2009)
QA Phi->KK (March 05 2009)
1. Dedx
Reconstruction on Kaon Daugthers. 2 different plots are shown just for the sake of completenees....
/Dedx_P08id_Phi_030509(Ghost.gif)
/Dedx_P08id_Phi_030509.gif)
2.Dca
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
/DCA_eta_pt_P08id_Phi_030509.gif)
3. Nfit
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
/Nfit_eta_pt_P08id_Phi_030509.gif)
4. Delta Vertex
/DeltaVertex_P08id_Phi_030509.gif)
5. Z Vertex and X vs Y vertex
/Vertex_P08id_Phi_030509.gif)
6. Global Variables : Phi and Rapidity
/Phi_P08id_Phi_030509.gif)
/Rapidity_P08id_Phi_030509.gif)
7. pt
/pT_P08id_Phi_030509.gif)
8. Randomness
/Y_pT_P08id_Phi_030509.gif)
/pT_Phi_P08id_Phi_030509.gif)
P08id (phi ->KK)
QA P08id (Phi->KK)
This is the QA for P08id (phi - > KK). econstructin on Global Tracks and just Kaons. Macro from Xianglei used (I found the QA macro very familiar). scale factor of 1.38 applied.
1. dEdx
Reconstruction on Kaon Daugthers. 2 different plots are shown just to see how Montecarlo looks on top of the Ghost Tracks
/dedx_f1_38(1).gif)
/dedx_v2_f1_38.gif)
2. DCA Distributions
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
/dca_eta_pt(1).gif)
3. NFit Distributions
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
/nfit_eta_pt(1).gif)
4. Delta Vertex
The following are the Delta Vertex ( Vertex Reconstructed - Vertex Embedded) plot for the 3 diiferent coordinates (x, y and z)
/DeltaVX.gif)
/DeltaVY.gif)
/DeltaVZ.gif)
5. Z Vertex and X vs Y vertex
/vertex_1_38.gif)
6. Global Variables : Phi and Rapidity
/phi_f1_38.gif)
/y_f1_38.gif)
7. Pt
Embedded Phi meson with flat pt (black)and Recosntructed Kaons (red).
/pt_f1_38.gif)
8. Randomness Plots
The following plots, are to check the randomness of the input Monte Carlo (MC) tracks.
/mc_pt_phi.gif)
/mc_pt_y.gif)
P08id(ALambda -> P, Pion)
QA ALambda->P, pi (03 08 2009)
1. Dedx
Reconstruction on Proton and Pion Daugthers. 2 different plots are shown just for the sake of completenees....
| Reco Proton | Reco Pion |
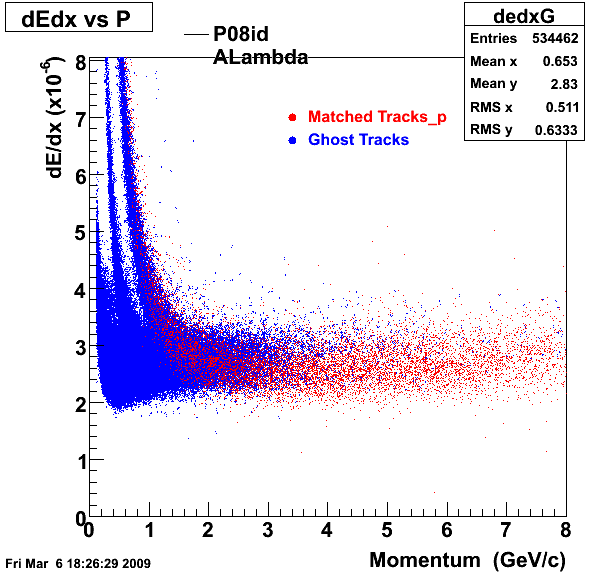 | 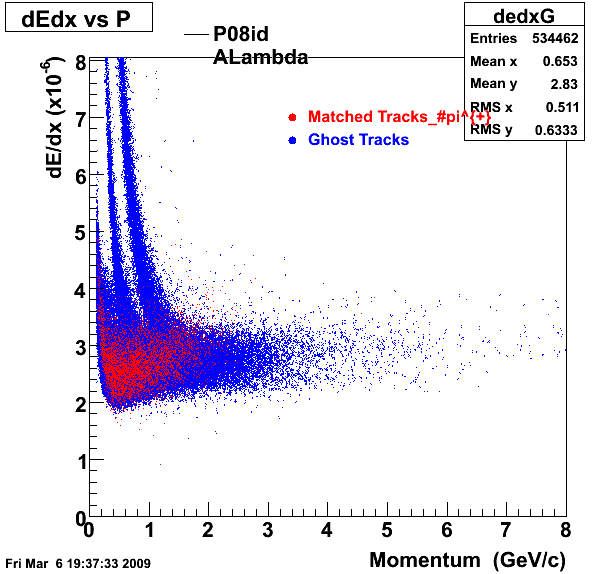 |
2.Dca
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
*.Reconstructing on Proton
*. Reconstructing on Pion
3. Nfit
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
*. Reconstructing Proton
*. Reconstructing Pion
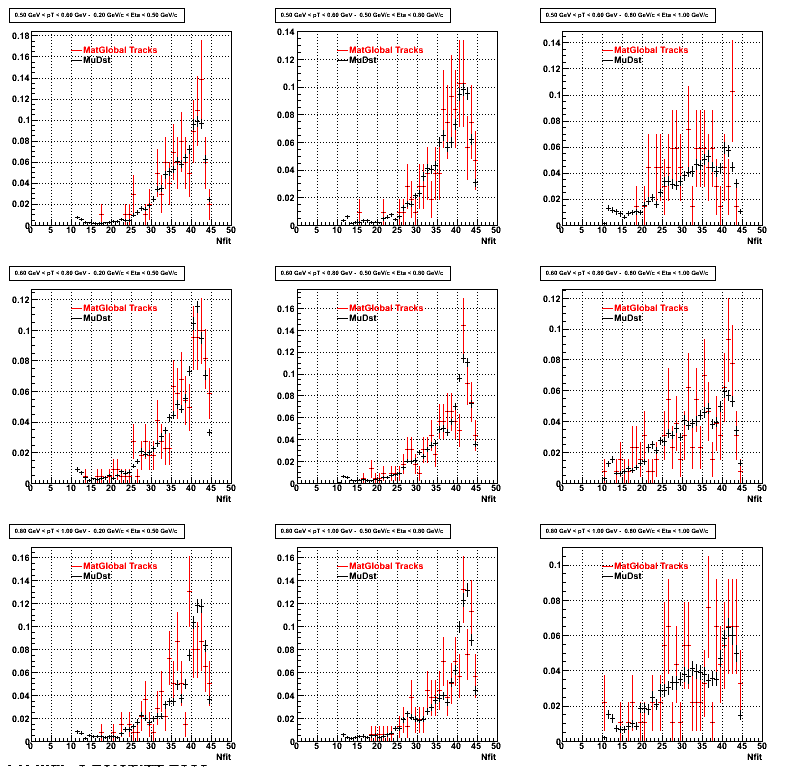
4. Delta Vertex
When reconstructed in Proton and pions it turns out to have the same ditreibutions so I just posted one of them
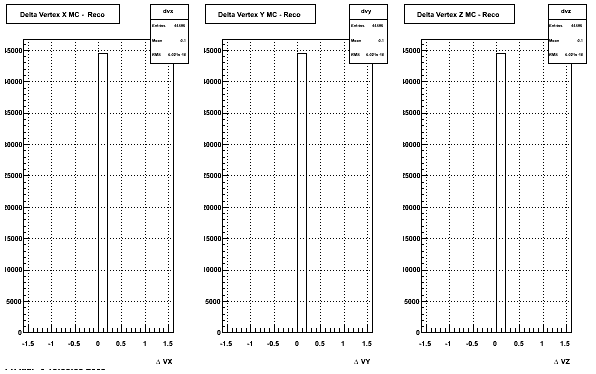
5. Z Vertex and X vs Y vertex
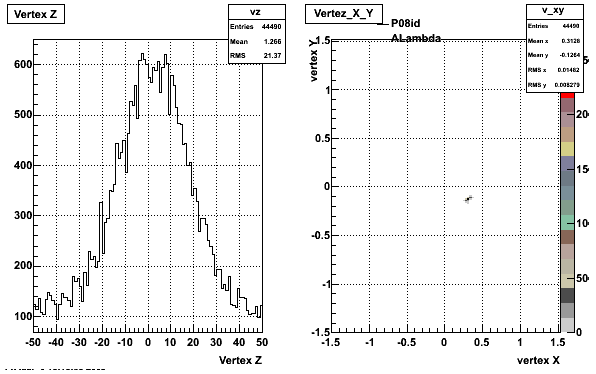
6. Global Variables : Phi and Rapidity
| Reco Proton | Reco Pion |
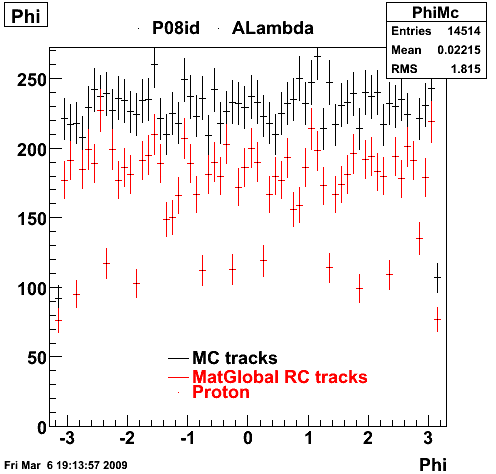 | 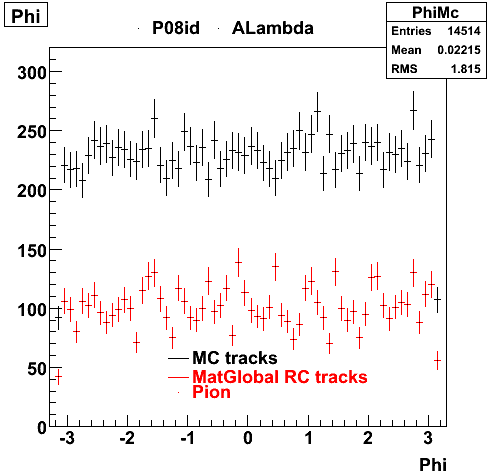 |
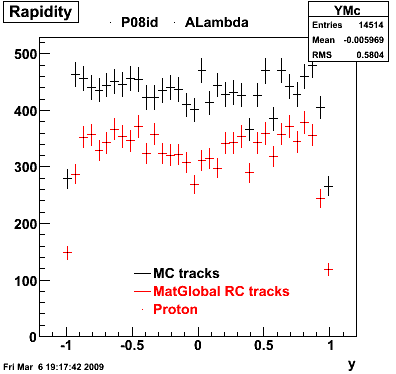 | 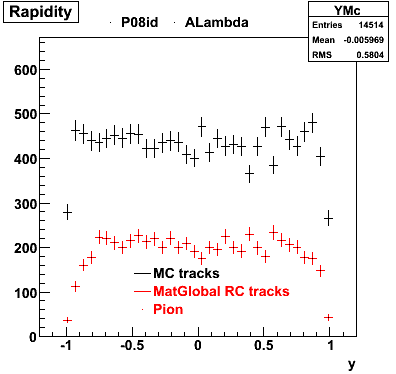 |
7. pt
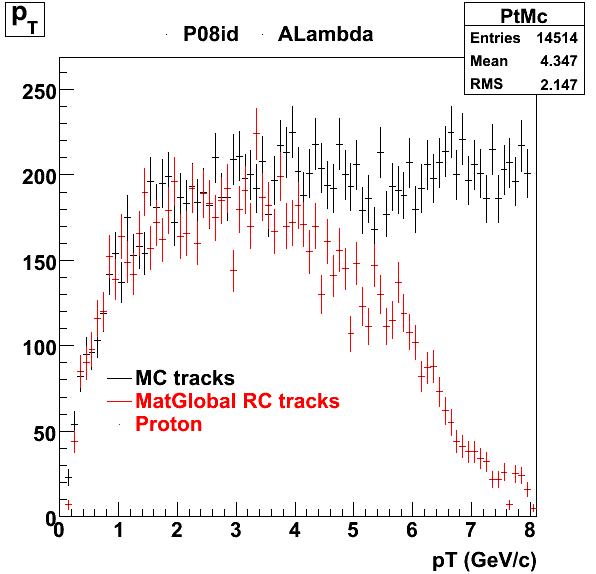
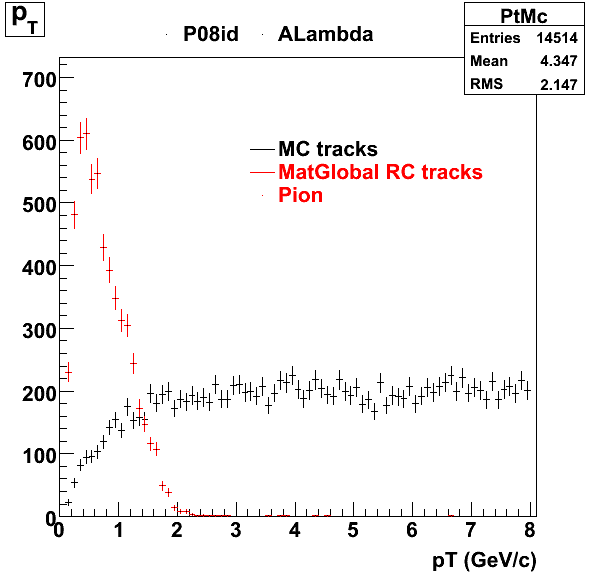
8. Randomness

P08id(Xi -> Reco(PiMinus)
Xi-> Lamba + Pion - ->P + Pion - + Pion -
(03 08 2009)
1. Dedx
Reconstruction on PI Minus

2.Dca
An original 3D histogram had been created and filled with pT, Eta and DCA as the 3 coordinates. Projection on PtBins and EtaBins had been made to create this "matrix" of plots.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
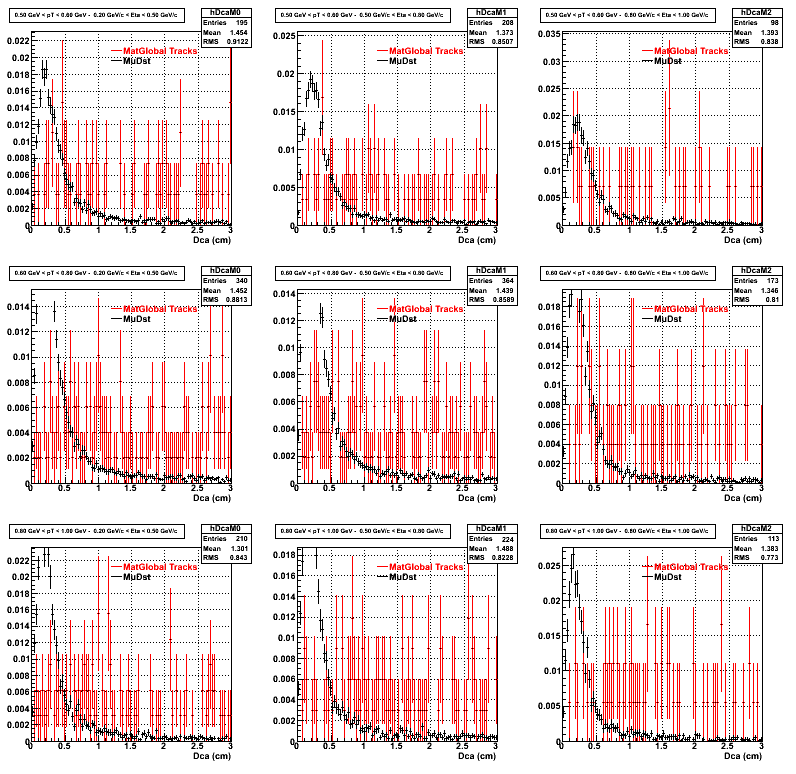
3. Nfit
Similarly An Original 3D Histogram had been created with Pt, Eta and Nfit as coordinates. Respective projections had been made in the same pT and Eta Bins as the DCa distributions.
Pt Bin array used : { 0.5, 0.6, 0.8, 1.0} (moves down) and
Eta Bin array : {0.2, 0.5, 0.8, 1.0} (moves to the right)
For the Error bars, i used the option hist->Sumw2();
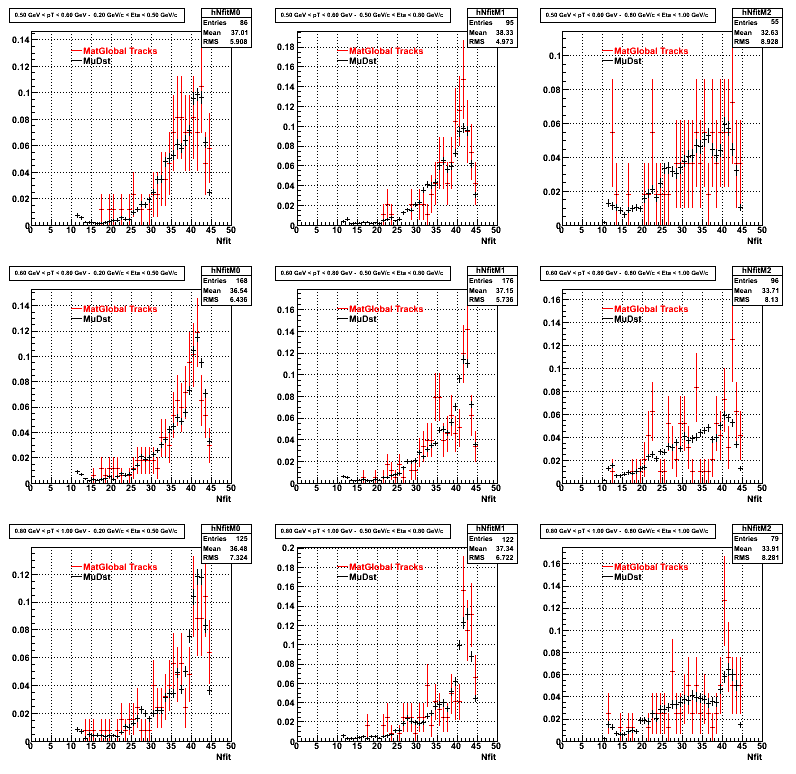
4. Delta Vertex

5. Z Vertex and X vs Y vertex
6. Global Variables : Phi and Rapidity

7. pt

8. Randomness
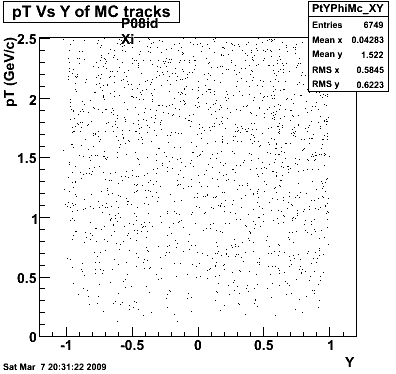
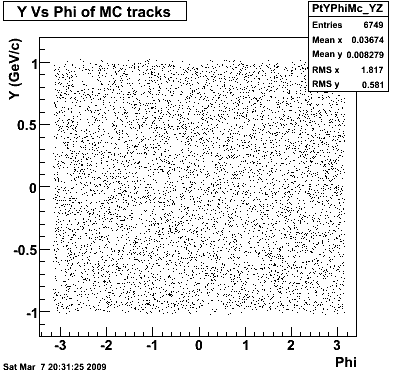
Phi->KK(Mar 12)
Please find the QA plots here
The data looks good.
Reconstructed phi meson has small rapidity dependence.
Yuri_Test_PionMinus_032009
![]() QA PionMinus
QA PionMinus

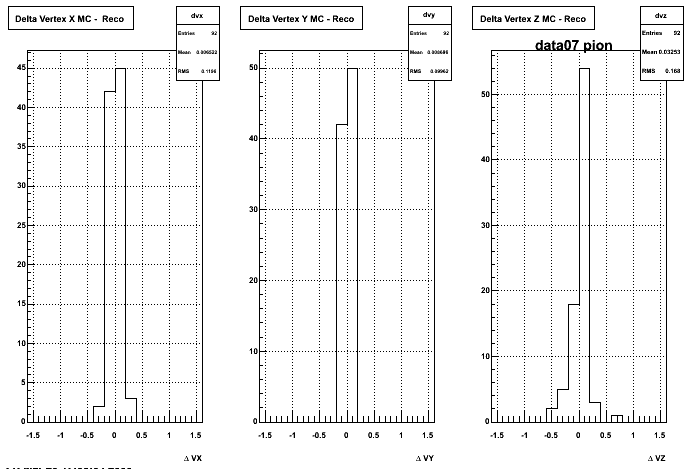
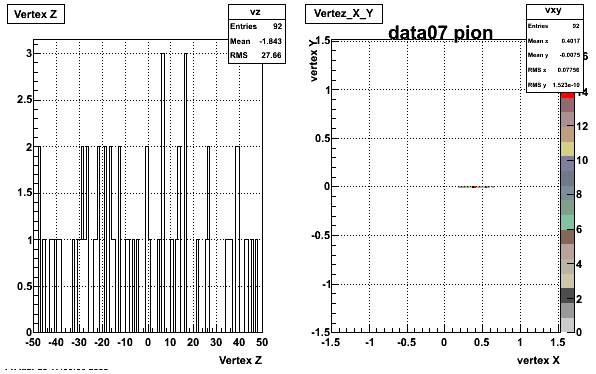
P04if
-
Hit level check-up:
Previous results - May 2002
This document is intended to provide all the information available for previous productions such that results for Quality Control studies and Productions Cross checks. Part of quality control studies include identification of missing/dead detector areas, distance of closest approach distributions (dca), and fit points distributions(Nfit). As part of production cross checks, some results in centrality dependance and efficiency are shown
Quality Control
These are dedx vs P graphs. All of them show reasonable agreement with data.(Done on May 2002)
| Pi Minus | K Minus | Proton |
| Pi Plus | K Plus | P Bar |
PI PLUS. In the following dca distributions some discrepancy is shown. Due to secondaries?
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | MinBias 0.2 GeV/c < pT < 0.3 Gev/c | Central 0.3 GeV/c <pT < 0.4 GeV/c |
PI MINUS. Some discrepancy is shown. Due to secondaries?
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | MinBias 0.2 GeV/c < pT < 0.3 Gev/c | Central 0.3 GeV/c <pT < 0.4 GeV/c |
K PLUS. In the following dca distributions Good agreement with data is shown.
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | MinBias 0.2 GeV/c < pT < 0.3 Gev/c | Central 0.3 GeV/c <pT < 0.4 GeV/c |
K MINUS. In the following dca distributions Good agreement with data is shown.
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | MinBias 0.2 GeV/c < pT < 0.3 Gev/c | Central 0.3 GeV/c <pT < 0.4 GeV/c |
PROTON. The real data Dca distribution is wider, especially at low pT -> Most likely due to secondary tracks in the sample. A tail from background protons dominating distribution at low pt can be clearly seen. Expected deviation from the primary MC tracks.
| Peripheral 0.2 GeV/c < pT < 0.3 Gev/c | MinBias 0.5 GeV/c < pT < 0.6 Gev/c | Central 0.7 GeV/c <pT < 0.8 GeV/c |
Pbar. The real data Dca distribution is wider, especially at low pT -> Most likely due to secondary tracks in the sample.
| Peripheral 0.2 GeV/c < pT < 0.3 Gev/c | MinBias 0.3 eV/c < pT < 0.4 Gev/c | Central 0.5 GeV/c <pT < 0.6 GeV/c |
PI PLUS. Good agreement with data is shown.
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | MinBias 0.2 GeV/c < pT < 0.3 Gev/c | Central 0.3 GeV/c <pT < 0.4 GeV/c |
PI MINUS. Good agreement with data is shown.
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | Peripheral 0.2 GeV/c < pT < 0.3 Gev/c | Peripheral 0.3 GeV/c < pT < 0.4 Gev/c |
K PLUS. Good agreement with data is shown.
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | MinBias 0.2 GeV/c < pT < 0.3 Gev/c | Central 0.3 GeV/c <pT < 0.4 GeV/c |
K MINUS. Good agreement with data is shown.
| Peripheral 0.1 GeV/c < pT < 0.2 Gev/c | Peripheral 0.2 GeV/c < pT < 0.3 Gev/c | Peripheral 0.3 GeV/c < pT < 0.4 Gev/c |
PROTON. Good agreement with data is shown.
| Peripheral 0.2 GeV/c < pT < 0.3 Gev/c | Peripheral 0.3 GeV/c < pT < 0.4 Gev/c | Peripheral 0.5 GeV/c < pT < 0.6 Gev/c |
Pbar. Good agreement with data is shown.
| Peripheral 0.2 GeV/c < pT < 0.3 Gev/c | Peripheral 0.3 GeV/c < pT < 0.4 Gev/c | Peripheral 0.5 GeV/c < pT < 0.6 Gev/c |